Targeting voltage sensors in sodium channels with spider toxins
- PMID: 20097434
- PMCID: PMC2847040
- DOI: 10.1016/j.tips.2009.12.007
Targeting voltage sensors in sodium channels with spider toxins
Abstract
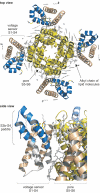
Voltage-activated sodium (Nav) channels are essential in generating and propagating nerve impulses, placing them amongst the most widely targeted ion channels by toxins from venomous organisms. An increasing number of spider toxins have been shown to interfere with the voltage-driven activation process of mammalian Nav channels, possibly by interacting with one or more of their voltage sensors. This review focuses on our existing knowledge of the mechanism by which spider toxins affect Nav channel gating and the possible applications of these toxins in the drug discovery process.
Figures


Similar articles
-
Spider-venom peptides that target voltage-gated sodium channels: pharmacological tools and potential therapeutic leads.Toxicon. 2012 Sep 15;60(4):478-91. doi: 10.1016/j.toxicon.2012.04.337. Epub 2012 Apr 20. Toxicon. 2012. PMID: 22543187 Review.
-
Insect-selective spider toxins targeting voltage-gated sodium channels.Toxicon. 2007 Mar 15;49(4):490-512. doi: 10.1016/j.toxicon.2006.11.027. Epub 2006 Dec 5. Toxicon. 2007. PMID: 17223149 Review.
-
Deconstructing voltage sensor function and pharmacology in sodium channels.Nature. 2008 Nov 13;456(7219):202-8. doi: 10.1038/nature07473. Nature. 2008. PMID: 19005548 Free PMC article.
-
Two tarantula peptides inhibit activation of multiple sodium channels.Biochemistry. 2002 Dec 17;41(50):14734-47. doi: 10.1021/bi026546a. Biochemistry. 2002. PMID: 12475222
-
Four novel tarantula toxins as selective modulators of voltage-gated sodium channel subtypes.Mol Pharmacol. 2006 Feb;69(2):419-29. doi: 10.1124/mol.105.015941. Epub 2005 Nov 2. Mol Pharmacol. 2006. PMID: 16267209
Cited by
-
Animal toxins influence voltage-gated sodium channel function.Handb Exp Pharmacol. 2014;221:203-29. doi: 10.1007/978-3-642-41588-3_10. Handb Exp Pharmacol. 2014. PMID: 24737238 Free PMC article. Review.
-
The tarantula toxin β/δ-TRTX-Pre1a highlights the importance of the S1-S2 voltage-sensor region for sodium channel subtype selectivity.Sci Rep. 2017 Apr 20;7(1):974. doi: 10.1038/s41598-017-01129-0. Sci Rep. 2017. PMID: 28428547 Free PMC article.
-
Optimizing Nav1.7-Targeted Analgesics: Revealing Off-Target Effects of Spider Venom-Derived Peptide Toxins and Engineering Strategies for Improvement.Adv Sci (Weinh). 2024 Nov;11(42):e2406656. doi: 10.1002/advs.202406656. Epub 2024 Sep 9. Adv Sci (Weinh). 2024. PMID: 39248322 Free PMC article.
-
Engineering Highly Potent and Selective Microproteins against Nav1.7 Sodium Channel for Treatment of Pain.J Biol Chem. 2016 Jul 1;291(27):13974-13986. doi: 10.1074/jbc.M116.725978. Epub 2016 Apr 22. J Biol Chem. 2016. PMID: 27129258 Free PMC article.
-
µ-Theraphotoxin Pn3a inhibition of CaV3.3 channels reveals a novel isoform-selective drug binding site.Elife. 2022 Jul 20;11:e74040. doi: 10.7554/eLife.74040. Elife. 2022. PMID: 35858123 Free PMC article.
References
-
- Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. - PubMed
-
- Hille B. Ion channels of excitable membranes. Sinauer Associates, Inc.; 2001.
-
- Cannon SC. Pathomechanisms in Channelopathies of Skeletal Muscle and Brain. Annu Rev Neurosci. 2006;29:387–415. - PubMed
-
- Goldin AL. Resurgence of sodium channel research. Annu Rev Physiol. 2001;63:871–894. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

