Loss of function of ATXN1 increases amyloid beta-protein levels by potentiating beta-secretase processing of beta-amyloid precursor protein
- PMID: 20097758
- PMCID: PMC2838273
- DOI: 10.1074/jbc.M109.079079
Loss of function of ATXN1 increases amyloid beta-protein levels by potentiating beta-secretase processing of beta-amyloid precursor protein
Abstract
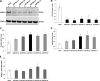
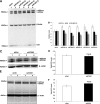
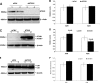
Alzheimer disease (AD) is a devastating neurodegenerative disease with complex and strong genetic inheritance. Four genes have been established to either cause familial early onset AD (APP, PSEN1, and PSEN2) or to increase susceptibility for late onset AD (APOE). To date approximately 80% of the late onset AD genetic variance remains elusive. Recently our genome-wide association screen identified four novel late onset AD candidate genes. Ataxin 1 (ATXN1) is one of these four AD candidate genes and has been indicated to be the disease gene for spinocerebellar ataxia type 1, which is also a neurodegenerative disease. Mounting evidence suggests that the excessive accumulation of Abeta, the proteolytic product of beta-amyloid precursor protein (APP), is the primary AD pathological event. In this study, we ask whether ATXN1 may lead to AD pathogenesis by affecting Abeta and APP processing utilizing RNA interference in a human neuronal cell model and mouse primary cortical neurons. We show that knock-down of ATXN1 significantly increases the levels of both Abeta40 and Abeta42. This effect could be rescued with concurrent overexpression of ATXN1. Moreover, overexpression of ATXN1 decreased Abeta levels. Regarding the underlying molecular mechanism, we show that the effect of ATXN1 expression on Abeta levels is modulated via beta-secretase cleavage of APP. Taken together, ATXN1 functions as a genetic risk modifier that contributes to AD pathogenesis through a loss-of-function mechanism by regulating beta-secretase cleavage of APP and Abeta levels.
Figures

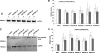

Similar articles
-
LRRTM3 promotes processing of amyloid-precursor protein by BACE1 and is a positional candidate gene for late-onset Alzheimer's disease.Proc Natl Acad Sci U S A. 2006 Nov 21;103(47):17967-72. doi: 10.1073/pnas.0605461103. Epub 2006 Nov 10. Proc Natl Acad Sci U S A. 2006. PMID: 17098871 Free PMC article.
-
RNA interference silencing of the adaptor molecules ShcC and Fe65 differentially affect amyloid precursor protein processing and Abeta generation.J Biol Chem. 2007 Feb 16;282(7):4318-4325. doi: 10.1074/jbc.M609293200. Epub 2006 Dec 14. J Biol Chem. 2007. PMID: 17170108
-
Relationship between ubiquilin-1 and BACE1 in human Alzheimer's disease and APdE9 transgenic mouse brain and cell-based models.Neurobiol Dis. 2016 Jan;85:187-205. doi: 10.1016/j.nbd.2015.11.005. Epub 2015 Nov 10. Neurobiol Dis. 2016. PMID: 26563932
-
Alzheimer's disease.Subcell Biochem. 2012;65:329-52. doi: 10.1007/978-94-007-5416-4_14. Subcell Biochem. 2012. PMID: 23225010 Review.
-
beta-Secretase, APP and Abeta in Alzheimer's disease.Subcell Biochem. 2005;38:79-103. Subcell Biochem. 2005. PMID: 15709474 Review.
Cited by
-
Integrated analysis of human genetic association study and mouse transcriptome suggests LBH and SHF genes as novel susceptible genes for amyloid-β accumulation in Alzheimer's disease.Hum Genet. 2018 Jul;137(6-7):521-533. doi: 10.1007/s00439-018-1906-z. Epub 2018 Jul 13. Hum Genet. 2018. PMID: 30006735 Free PMC article.
-
Degradation and inhibition of epigenetic regulatory protein BRD4 exacerbate Alzheimer's disease-related neuropathology in cell models.J Biol Chem. 2022 Apr;298(4):101794. doi: 10.1016/j.jbc.2022.101794. Epub 2022 Mar 3. J Biol Chem. 2022. PMID: 35248531 Free PMC article.
-
Amyloid-β production via cleavage of amyloid-β protein precursor is modulated by cell density.J Alzheimers Dis. 2010;22(2):683-984. doi: 10.3233/JAD-2010-100816. J Alzheimers Dis. 2010. PMID: 20847415 Free PMC article.
-
Concerted perturbation observed in a hub network in Alzheimer's disease.PLoS One. 2012;7(7):e40498. doi: 10.1371/journal.pone.0040498. Epub 2012 Jul 16. PLoS One. 2012. PMID: 22815752 Free PMC article.
-
Curcumin decreases amyloid-beta peptide levels by attenuating the maturation of amyloid-beta precursor protein.J Biol Chem. 2010 Sep 10;285(37):28472-80. doi: 10.1074/jbc.M110.133520. Epub 2010 Jul 9. J Biol Chem. 2010. PMID: 20622013 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

