Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants: characterization of ABSCISIC ACID INSENSITIVE1-like type 2C protein phosphatase in the liverwort Marchantia polymorpha
- PMID: 20097789
- PMCID: PMC2832234
- DOI: 10.1104/pp.110.153387
Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants: characterization of ABSCISIC ACID INSENSITIVE1-like type 2C protein phosphatase in the liverwort Marchantia polymorpha
Abstract
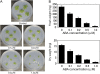
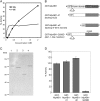
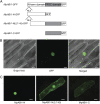
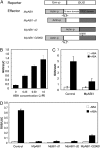
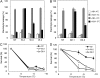
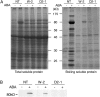
Abscisic acid (ABA) is postulated to be a ubiquitous hormone that plays a central role in seed development and responses to environmental stresses of vascular plants. However, in liverworts (Marchantiophyta), which represent the oldest extant lineage of land plants, the role of ABA has been least emphasized; thus, very little information is available on the molecular mechanisms underlying ABA responses. In this study, we isolated and characterized MpABI1, an ortholog of ABSCISIC ACID INSENSITIVE1 (ABI1), from the liverwort Marchantia polymorpha. The MpABI1 cDNA encoded a 568-amino acid protein consisting of the carboxy-terminal protein phosphatase 2C (PP2C) domain and a novel amino-terminal regulatory domain. The MpABI1 transcript was detected in the gametophyte, and its expression level was increased by exogenous ABA treatment in the gemma, whose growth was strongly inhibited by ABA. Experiments using green fluorescent protein fusion constructs indicated that MpABI1 was mainly localized in the nucleus and that its nuclear localization was directed by the amino-terminal domain. Transient overexpression of MpABI1 in M. polymorpha and Physcomitrella patens cells resulted in suppression of ABA-induced expression of the wheat Em promoter fused to the beta -glucuronidase gene. Transgenic P. patens expressing MpABI1 and its mutant construct, MpABI1-d2, lacking the amino-terminal domain, had reduced freezing and osmotic stress tolerance, and associated with reduced accumulation of ABA-induced late embryogenesis abundant-like boiling-soluble proteins. Furthermore, ABA-induced morphological changes leading to brood cells were not prominent in these transgenic plants. These results suggest that MpABI1 is a negative regulator of ABA signaling, providing unequivocal molecular evidence of PP2C-mediated ABA response mechanisms functioning in liverworts.
Figures




Similar articles
-
Abscisic acid induces biosynthesis of bisbibenzyls and tolerance to UV-C in the liverwort Marchantia polymorpha.Phytochemistry. 2015 Sep;117:547-553. doi: 10.1016/j.phytochem.2015.05.009. Epub 2015 Jun 5. Phytochemistry. 2015. PMID: 26055979
-
Abscisic acid-induced gene expression in the liverwort Marchantia polymorpha is mediated by evolutionarily conserved promoter elements.Physiol Plant. 2016 Apr;156(4):407-20. doi: 10.1111/ppl.12385. Epub 2015 Oct 12. Physiol Plant. 2016. PMID: 26456006
-
Functional analyses of the ABI1-related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens.Plant Mol Biol. 2009 Jun;70(3):327-40. doi: 10.1007/s11103-009-9476-z. Epub 2009 Mar 6. Plant Mol Biol. 2009. PMID: 19266168
-
Abscisic acid and abiotic stress tolerance - different tiers of regulation.J Plant Physiol. 2014 Apr 15;171(7):486-96. doi: 10.1016/j.jplph.2013.12.007. J Plant Physiol. 2014. PMID: 24655384 Review.
-
Thirsty plants and beyond: structural mechanisms of abscisic acid perception and signaling.Curr Opin Struct Biol. 2010 Dec;20(6):722-9. doi: 10.1016/j.sbi.2010.09.007. Epub 2010 Oct 14. Curr Opin Struct Biol. 2010. PMID: 20951573 Free PMC article. Review.
Cited by
-
Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins.J Biol Chem. 2012 Mar 9;287(11):7956-68. doi: 10.1074/jbc.M111.279331. Epub 2012 Jan 17. J Biol Chem. 2012. PMID: 22253446 Free PMC article.
-
Isolation and functional characterization of the promoter of a DEAD-box helicase Psp68 using Agrobacterium-mediated transient assay.Plant Signal Behav. 2014 Apr 30;9:e28992. doi: 10.4161/psb.28992. Online ahead of print. Plant Signal Behav. 2014. PMID: 24785194 Free PMC article.
-
Cryopreservation of Gemmae from the Liverwort Marchantia polymorpha L.Plant Cell Physiol. 2016 Feb;57(2):300-6. doi: 10.1093/pcp/pcv173. Epub 2015 Nov 11. Plant Cell Physiol. 2016. PMID: 26561534 Free PMC article.
-
Environmental Pollutant Anthracene Induces ABA-Dependent Transgenerational Effects on Gemmae Dormancy in Marchantia polymorpha.Plants (Basel). 2024 Oct 25;13(21):2979. doi: 10.3390/plants13212979. Plants (Basel). 2024. PMID: 39519898 Free PMC article.
-
Differential regulations of abscisic acid-induced desiccation tolerance and vegetative dormancy by group B3 Raf kinases in liverworts.Front Plant Sci. 2022 Jul 28;13:952820. doi: 10.3389/fpls.2022.952820. eCollection 2022. Front Plant Sci. 2022. PMID: 35968153 Free PMC article.
References
-
- Attree SM, Pomeroy MK, Fowke LC. (1995) Development of white spruce (Picea glauca (Moench.) Voss) somatic embryos during culture with abscisic acid and osmoticum, and their tolerance to drying and frozen storage. J Exp Bot 46: 433–439
-
- Bertauche N, Leung J, Giraudat J. (1996) Protein phosphatase activity of abscisic acid insensitive I (ABII) protein from Arabidopsis thaliana. Eur J Biochem 241: 193–200 - PubMed
-
- Bowman JL, Floyd SK, Sakakibara K. (2007) Green genes: comparative genomics of the green branch of life. Cell 129: 229–234 - PubMed
-
- Chandler PM, Robertson M. (1994) Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu Rev Plant Physiol Plant Mol Biol 45: 113–141
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

