Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii
- PMID: 20097827
- PMCID: PMC2838008
- DOI: 10.1128/AEM.02670-09
Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii
Abstract
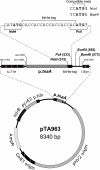
Research into archaea will not achieve its full potential until systems are in place to carry out genetics and biochemistry in the same species. Haloferax volcanii is widely regarded as the best-equipped organism for archaeal genetics, but the development of tools for the expression and purification of H. volcanii proteins has been neglected. We have developed a series of plasmid vectors and host strains for conditional overexpression of halophilic proteins in H. volcanii. The plasmids feature the tryptophan-inducible p.tnaA promoter and a 6xHis tag for protein purification by metal affinity chromatography. Purification is facilitated by host strains, where pitA is replaced by the ortholog from Natronomonas pharaonis. The latter lacks the histidine-rich linker region found in H. volcanii PitA and does not copurify with His-tagged recombinant proteins. We also deleted the mrr restriction endonuclease gene, thereby allowing direct transformation without the need to passage DNA through an Escherichia coli dam mutant.
Figures


Similar articles
-
Isolation of fusion proteins containing SecY and SecE, components of the protein translocation complex from the halophilic archaeon Haloferax volcanii.Extremophiles. 2003 Feb;7(1):71-7. doi: 10.1007/s00792-002-0297-0. Epub 2002 Oct 3. Extremophiles. 2003. PMID: 12579382
-
Towards a systems approach in the genetic analysis of archaea: Accelerating mutant construction and phenotypic analysis in Haloferax volcanii.Archaea. 2010 Dec 23;2010:426239. doi: 10.1155/2010/426239. Archaea. 2010. PMID: 21234384 Free PMC article.
-
Characterization of a tightly controlled promoter of the halophilic archaeon Haloferax volcanii and its use in the analysis of the essential cct1 gene.Mol Microbiol. 2007 Dec;66(5):1092-106. doi: 10.1111/j.1365-2958.2007.05980.x. Epub 2007 Oct 31. Mol Microbiol. 2007. PMID: 17973910
-
Haloferax volcanii.Trends Microbiol. 2019 Jan;27(1):86-87. doi: 10.1016/j.tim.2018.10.004. Epub 2018 Nov 17. Trends Microbiol. 2019. PMID: 30459094 Review.
-
Haloferax volcanii for biotechnology applications: challenges, current state and perspectives.Appl Microbiol Biotechnol. 2020 Feb;104(4):1371-1382. doi: 10.1007/s00253-019-10314-2. Epub 2019 Dec 20. Appl Microbiol Biotechnol. 2020. PMID: 31863144 Free PMC article. Review.
Cited by
-
Expression in Haloferax volcanii of 3-hydroxy-3-methylglutaryl coenzyme A synthase facilitates isolation and characterization of the active form of a key enzyme required for polyisoprenoid cell membrane biosynthesis in halophilic archaea.J Bacteriol. 2013 Sep;195(17):3854-62. doi: 10.1128/JB.00485-13. Epub 2013 Jun 21. J Bacteriol. 2013. PMID: 23794621 Free PMC article.
-
d-Ribose Catabolism in Archaea: Discovery of a Novel Oxidative Pathway in Haloarcula Species.J Bacteriol. 2020 Jan 15;202(3):e00608-19. doi: 10.1128/JB.00608-19. Print 2020 Jan 15. J Bacteriol. 2020. PMID: 31712277 Free PMC article.
-
A sweet new set of inducible and constitutive promoters in Haloferax volcanii.Front Microbiol. 2023 Aug 10;14:1204876. doi: 10.3389/fmicb.2023.1204876. eCollection 2023. Front Microbiol. 2023. PMID: 37637112 Free PMC article.
-
A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii.J Biol Chem. 2014 Mar 7;289(10):7164-7177. doi: 10.1074/jbc.M113.508184. Epub 2014 Jan 23. J Biol Chem. 2014. PMID: 24459147 Free PMC article.
-
Division plane placement in pleomorphic archaea is dynamically coupled to cell shape.Mol Microbiol. 2019 Sep;112(3):785-799. doi: 10.1111/mmi.14316. Epub 2019 Jun 11. Mol Microbiol. 2019. PMID: 31136034 Free PMC article.
References
-
- Aertsen, A., M. Tesfazgi Mebrhatu, and C. W. Michiels. 2008. Activation of the Salmonella typhimurium Mrr protein. Biochem. Biophys. Res. Commun. 367:435-439. - PubMed
-
- Allers, T., and M. Mevarech. 2005. Archaeal genetics—the third way. Nat. Rev. Genet. 6:58-73. - PubMed
-
- Bab-Dinitz, E., H. Shmuely, J. Maupin-Furlow, J. Eichler, and B. Shaanan. 2006. Haloferax volcanii PitA: an example of functional interaction between the Pfam chlorite dismutase and antibiotic biosynthesis monooxygenase families? Bioinformatics 22:671-675. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

