Precise excision of IS5 from the intergenic region between the fucPIK and the fucAO operons and mutational control of fucPIK operon expression in Escherichia coli
- PMID: 20097855
- PMCID: PMC2838058
- DOI: 10.1128/JB.01085-09
Precise excision of IS5 from the intergenic region between the fucPIK and the fucAO operons and mutational control of fucPIK operon expression in Escherichia coli
Abstract
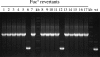
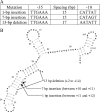
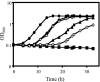
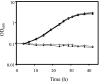
Excision of transposable genetic elements from host DNA occurs at low frequencies and is usually imprecise. A common insertion sequence element in Escherichia coli, IS5, has been shown to provide various benefits to its host by inserting into specific sites. Precise excision of this element had not previously been demonstrated. Using a unique system, the fucose (fuc) regulon, in which IS5 insertion and excision result in two distinct selectable phenotypes, we have demonstrated that IS5 can precisely excise from its insertion site, restoring the wild-type phenotype. In addition to precise excision, several "suppressor" insertion, deletion, and point mutations restore the wild-type Fuc(+) phenotype to various degrees without IS5 excision. The possible bases for these observations are discussed.
Figures

Similar articles
-
Constitutive activation of the fucAO operon and silencing of the divergently transcribed fucPIK operon by an IS5 element in Escherichia coli mutants selected for growth on L-1,2-propanediol.J Bacteriol. 1989 Nov;171(11):6097-105. doi: 10.1128/jb.171.11.6097-6105.1989. J Bacteriol. 1989. PMID: 2553671 Free PMC article.
-
A cyclic AMP receptor protein mutant that constitutively activates an Escherichia coli promoter disrupted by an IS5 insertion.J Bacteriol. 1999 Dec;181(24):7457-63. doi: 10.1128/JB.181.24.7457-7463.1999. J Bacteriol. 1999. PMID: 10601201 Free PMC article.
-
Transcriptional mechanism by which IS5 activates the fucAO operon in Escherichia coli.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf172. doi: 10.1093/nar/gkaf172. Nucleic Acids Res. 2025. PMID: 40066878 Free PMC article.
-
Transposon-mediated directed mutation in bacteria and eukaryotes.Front Biosci (Landmark Ed). 2017 Mar 1;22(9):1458-1468. doi: 10.2741/4553. Front Biosci (Landmark Ed). 2017. PMID: 28199212 Free PMC article. Review.
-
Transposon-mediated adaptive and directed mutations and their potential evolutionary benefits.J Mol Microbiol Biotechnol. 2011;21(1-2):59-70. doi: 10.1159/000333108. Epub 2012 Jan 13. J Mol Microbiol Biotechnol. 2011. PMID: 22248543 Free PMC article. Review.
Cited by
-
Insertion Sequence (IS) Element-Mediated Activating Mutations of the Cryptic Aromatic β-Glucoside Utilization (BglGFB) Operon Are Promoted by the Anti-Terminator Protein (BglG) in Escherichia coli.Int J Mol Sci. 2022 Jan 28;23(3):1505. doi: 10.3390/ijms23031505. Int J Mol Sci. 2022. PMID: 35163427 Free PMC article.
-
Investigating How Genomic Contexts Impact IS5 Transposition Within the Escherichia coli Genome.Microorganisms. 2024 Dec 16;12(12):2600. doi: 10.3390/microorganisms12122600. Microorganisms. 2024. PMID: 39770802 Free PMC article.
-
Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness.Nature. 2014 Oct 30;514(7524):638-41. doi: 10.1038/nature13823. Epub 2014 Oct 1. Nature. 2014. PMID: 25274297 Free PMC article.
-
Hopping into a hot seat: Role of DNA structural features on IS5-mediated gene activation and inactivation under stress.PLoS One. 2017 Jun 30;12(6):e0180156. doi: 10.1371/journal.pone.0180156. eCollection 2017. PLoS One. 2017. PMID: 28666002 Free PMC article.
-
Insertion sequence-driven evolution of Escherichia coli in chemostats.J Mol Evol. 2011 Apr;72(4):398-412. doi: 10.1007/s00239-011-9439-2. Epub 2011 Mar 12. J Mol Evol. 2011. PMID: 21399911
References
-
- Ahmed, A. 1986. Evidence for replicative transposition of Tn5 and Tn9. J. Mol. Biol. 191:75-84. - PubMed
-
- Berg, D. E. 1977. Insertion and excision of the transposable kanamycin resisrance determinant Tn5, p. 205-212. In A. I. Bukhari, J. A. Shapiro, and S. L. Adhya (ed.), DNA insertion element, plasmids and episomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
-
- Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

