Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic States in Arabidopsis
- PMID: 20097869
- PMCID: PMC2828703
- DOI: 10.1105/tpc.109.072819
Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic States in Arabidopsis
Abstract
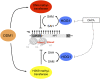
Epigenetic changes of gene expression can potentially be reversed by developmental programs, genetic manipulation, or pharmacological interference. However, a case of transcriptional gene silencing, originally observed in tetraploid Arabidopsis thaliana plants, created an epiallele resistant to many mutations or inhibitor treatments that activate many other suppressed genes. This raised the question about the molecular basis of this extreme stability. A combination of forward and reverse genetics and drug application provides evidence for an epigenetic double lock that is only alleviated upon the simultaneous removal of both DNA methylation and histone methylation. Therefore, the cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic states and contributes to heritable diversity of gene expression patterns.
Figures
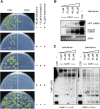

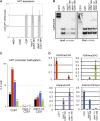

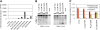
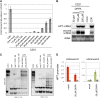
References
-
- Adams K.L., Wendel J.F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141 - PubMed
-
- Baubec T. (2008). Maintenance of polyploidy-associated transcriptional gene silencing in Arabidopsis thaliana, dissertation (Austria: University of Vienna). http://othes.univie.ac.at/3756/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

