MRL1, a conserved Pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis
- PMID: 20097872
- PMCID: PMC2828700
- DOI: 10.1105/tpc.109.066266
MRL1, a conserved Pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis
Abstract
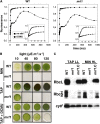
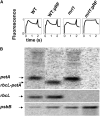
We identify and functionally characterize MRL1, a conserved nuclear-encoded regulator of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. The nonphotosynthetic mrl1 mutant of Chlamydomonas reinhardtii lacks ribulose-1,5-bisphosphate carboxylase/oxygenase, and the resulting block in electron transfer is partially compensated by redirecting electrons toward molecular oxygen via the Mehler reaction. This allows continued electron flow and constitutive nonphotochemical quenching, enhancing cell survival during illumination in spite of photosystem II and photosystem I photoinhibition. The mrl1 mutant transcribes rbcL normally, but the mRNA is unstable. The molecular target of MRL1 is the 5 ' untranslated region of rbcL. MRL1 is located in the chloroplast stroma, in a high molecular mass complex. Treatment with RNase or deletion of the rbcL gene induces a shift of the complex toward lower molecular mass fractions. MRL1 is well conserved throughout the green lineage, much more so than the 10 other pentatricopeptide repeat proteins found in Chlamydomonas. Depending upon the organism, MRL1 contains 11 to 14 pentatricopeptide repeats followed by a novel MRL1-C domain. In Arabidopsis thaliana, MRL1 also acts on rbcL and is necessary for the production/stabilization of the processed transcript, presumably because it acts as a barrier to 5 ' >3 ' degradation. The Arabidopsis mrl1 mutant retains normal levels of the primary transcript and full photosynthetic capacity.
Figures







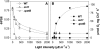

Similar articles
-
The chloroplast RNA-binding protein CP29A supports rbcL expression during cold acclimation.Proc Natl Acad Sci U S A. 2025 Feb 4;122(5):e2403969122. doi: 10.1073/pnas.2403969122. Epub 2025 Jan 29. Proc Natl Acad Sci U S A. 2025. PMID: 39879235 Free PMC article.
-
Manipulating RuBisCO accumulation in the green alga, Chlamydomonas reinhardtii.Plant Mol Biol. 2011 Jul;76(3-5):397-405. doi: 10.1007/s11103-011-9783-z. Epub 2011 May 24. Plant Mol Biol. 2011. PMID: 21607658
-
Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts.Mol Cell Biol. 2008 Sep;28(17):5529-42. doi: 10.1128/MCB.02056-07. Epub 2008 Jun 23. Mol Cell Biol. 2008. PMID: 18573878 Free PMC article.
-
Functional genomics of plant photosynthesis in the fast lane using Chlamydomonas reinhardtii.Trends Plant Sci. 2001 Aug;6(8):364-71. doi: 10.1016/s1360-1385(01)02018-0. Trends Plant Sci. 2001. PMID: 11495790 Review.
-
Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase.Arch Biochem Biophys. 2003 Jun 15;414(2):141-9. doi: 10.1016/s0003-9861(03)00171-1. Arch Biochem Biophys. 2003. PMID: 12781765 Review.
Cited by
-
The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria.Nucleic Acids Res. 2013 Jul;41(13):6650-63. doi: 10.1093/nar/gkt337. Epub 2013 May 8. Nucleic Acids Res. 2013. PMID: 23658225 Free PMC article.
-
Systematic sequencing of chloroplast transcript termini from Arabidopsis thaliana reveals >200 transcription initiation sites and the extensive imprints of RNA-binding proteins and secondary structures.Nucleic Acids Res. 2019 Dec 16;47(22):11889-11905. doi: 10.1093/nar/gkz1059. Nucleic Acids Res. 2019. PMID: 31732725 Free PMC article.
-
Novel shuttle markers for nuclear transformation of the green alga Chlamydomonas reinhardtii.Eukaryot Cell. 2011 Dec;10(12):1670-8. doi: 10.1128/EC.05043-11. Epub 2011 Oct 14. Eukaryot Cell. 2011. PMID: 22002656 Free PMC article.
-
Silencing of AtRAP, a target gene of a bacteria-induced small RNA, triggers antibacterial defense responses through activation of LSU2 and down-regulation of GLK1.New Phytol. 2017 Aug;215(3):1144-1155. doi: 10.1111/nph.14654. Epub 2017 Jun 28. New Phytol. 2017. PMID: 28656601 Free PMC article.
-
Can phenotypic plasticity in Rubisco performance contribute to photosynthetic acclimation?Photosynth Res. 2014 Feb;119(1-2):203-14. doi: 10.1007/s11120-013-9816-3. Epub 2013 Mar 31. Photosynth Res. 2014. PMID: 23543330 Review.
References
-
- Alonso J.M., Stepanova A.N. (2003). T-DNA mutagenesis in Arabidopsis. Methods Mol. Biol. 236: 177–188 - PubMed
-
- Anisimova M., Gascuel O. (2006). Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 55: 539–552 - PubMed
-
- Barkan A., Goldschmidt-Clermont M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

