Iminodiacetic-phosphoramidates as metabolic prototypes for diversifying nucleic acid polymerization in vivo
- PMID: 20097909
- PMCID: PMC2860114
- DOI: 10.1093/nar/gkp1246
Iminodiacetic-phosphoramidates as metabolic prototypes for diversifying nucleic acid polymerization in vivo
Abstract
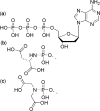
Previous studies in our laboratory proved that certain functional groups are able to mimic the pyrophosphate moiety and act as leaving groups in the enzymatic polymerization of deoxyribonucleic acids by HIV-1 reverse transcriptase. When the potential leaving group possesses two carboxylic acid moieties linked to the nucleoside via a phosphoramidate bond, it is efficiently recognized by this error-prone enzyme, resulting in nucleotide incorporation into DNA. Here, we present a new efficient alternative leaving group, iminodiacetic acid, which displays enhanced kinetics and an enhanced elongation capacity compared to previous results obtained with amino acid deoxyadenosine phosphoramidates. Iminodiacetic acid phosphoramidate of deoxyadenosine monophosphate (IDA-dAMP) is processed by HIV-1 RT as a substrate for single nucleotide incorporation and displays a typical Michaelis-Menten kinetic profile. This novel substrate also proved to be successful in primer strand elongation of a seven-base template overhang. Modelling of this new substrate in the active site of the enzyme revealed that the interactions formed between the triphosphate moiety, magnesium ions and enzyme's residues could be different from those of the natural triphosphate substrate and is likely to involve additional amino acid residues. Preliminary testing for a potential metabolic accessibility lets us to envision its possible use in an orthogonal system for nucleic acid synthesis that would not influence or be influenced by genetic information from the outside.
Figures
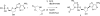
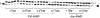

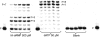

References
-
- Herdewijn P, Marliere P. Toward safe genetically modified organisms through the chemical diversification of nucleic acids. Chem. Biodivers. 2009;6:791–808. - PubMed
-
- Ghadessy FJ, Ramsay N, Boudsocq F, Loakes D, Brown A, Iwai S, Vaisman A, Woodgate R, Holliger P. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nat. Biotechnol. 2004;22:755–759. - PubMed
-
- Pochet S, Kaminski A, Van Aerschot A, Herdewijn P, Marlière P. Replication of hexitol oligonucleotides as a prelude to the propagation of a third type of nucleic acid in vivo. C.R. Biol. 2003;326:1175–1184. - PubMed
-
- Renders M, Lievrouw R, Krecmerova M, Holy A, Herdewijn P. Enzymatic polymerization of phosphonate nucleosides. Chembiochem. 2008;9:2883–2888. - PubMed
-
- Adelfinskaya O, Herdewijn P. Amino acid phosphoramidate nucleotides as alternative substrates for HIV-1 reverse transcriptase. Angew. Chem. Int. Ed. 2007;46:4356–4358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

