Sphingosine 1-phosphate lyase deficiency disrupts lipid homeostasis in liver
- PMID: 20097939
- PMCID: PMC2856294
- DOI: 10.1074/jbc.M109.081489
Sphingosine 1-phosphate lyase deficiency disrupts lipid homeostasis in liver
Abstract
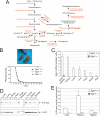
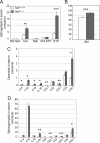
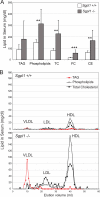
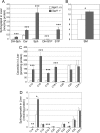
The cleavage of sphingoid base phosphates by sphingosine-1-phosphate (S1P) lyase to produce phosphoethanolamine and a fatty aldehyde is the final degradative step in the sphingolipid metabolic pathway. We have studied mice with an inactive S1P lyase gene and have found that, in addition to the expected increase of sphingoid base phosphates, other sphingolipids (including sphingosine, ceramide, and sphingomyelin) were substantially elevated in the serum and/or liver of these mice. This latter increase is consistent with a reutilization of the sphingosine backbone for sphingolipid synthesis due to its inability to exit the sphingolipid metabolic pathway. Furthermore, the S1P lyase deficiency resulted in changes in the levels of serum and liver lipids not directly within the sphingolipid pathway, including phospholipids, triacyglycerol, diacylglycerol, and cholesterol. Even though lipids in serum and lipid storage were elevated in liver, adiposity was reduced in the S1P lyase-deficient mice. Microarray analysis of lipid metabolism genes in liver showed that the S1P lyase deficiency caused widespread changes in their expression pattern, with a significant increase in the expression of PPARgamma, a master transcriptional regulator of lipid metabolism. However, the mRNA expression of the genes encoding the sphingosine kinases and S1P phosphatases, which directly control the levels of S1P, were not significantly changed in liver of the S1P lyase-deficient mice. These results demonstrate that S1P lyase is a key regulator of the levels of multiple sphingolipid substrates and reveal functional links between the sphingolipid metabolic pathway and other lipid metabolic pathways that may be mediated by shared lipid substrates and changes in gene expression programs. The disturbance of lipid homeostasis by altered sphingolipid levels may be relevant to metabolic diseases.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

