Application of gene network analysis techniques identifies AXIN1/PDIA2 and endoglin haplotypes associated with bicuspid aortic valve
- PMID: 20098615
- PMCID: PMC2809109
- DOI: 10.1371/journal.pone.0008830
Application of gene network analysis techniques identifies AXIN1/PDIA2 and endoglin haplotypes associated with bicuspid aortic valve
Abstract
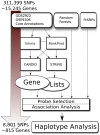
Bicuspid Aortic Valve (BAV) is a highly heritable congenital heart defect. The low frequency of BAV (1% of general population) limits our ability to perform genome-wide association studies. We present the application of four a priori SNP selection techniques, reducing the multiple-testing penalty by restricting analysis to SNPs relevant to BAV in a genome-wide SNP dataset from a cohort of 68 BAV probands and 830 control subjects. Two knowledge-based approaches, CANDID and STRING, were used to systematically identify BAV genes, and their SNPs, from the published literature, microarray expression studies and a genome scan. We additionally tested Functionally Interpolating SNPs (fitSNPs) present on the array; the fourth consisted of SNPs selected by Random Forests, a machine learning approach. These approaches reduced the multiple testing penalty by lowering the fraction of the genome probed to 0.19% of the total, while increasing the likelihood of studying SNPs within relevant BAV genes and pathways. Three loci were identified by CANDID, STRING, and fitSNPS. A haplotype within the AXIN1-PDIA2 locus (p-value of 2.926x10(-06)) and a haplotype within the Endoglin gene (p-value of 5.881x10(-04)) were found to be strongly associated with BAV. The Random Forests approach identified a SNP on chromosome 3 in association with BAV (p-value 5.061x10(-06)). The results presented here support an important role for genetic variants in BAV and provide support for additional studies in well-powered cohorts. Further, these studies demonstrate that leveraging existing expression and genomic data in the context of GWAS studies can identify biologically relevant genes and pathways associated with a congenital heart defect.
Conflict of interest statement
Figures





Similar articles
-
Defects in the Exocyst-Cilia Machinery Cause Bicuspid Aortic Valve Disease and Aortic Stenosis.Circulation. 2019 Oct 15;140(16):1331-1341. doi: 10.1161/CIRCULATIONAHA.119.038376. Epub 2019 Aug 7. Circulation. 2019. PMID: 31387361 Free PMC article.
-
Identification of Gender-Specific Genetic Variants in Patients With Bicuspid Aortic Valve.Am J Cardiol. 2016 Feb 1;117(3):420-6. doi: 10.1016/j.amjcard.2015.10.058. Epub 2015 Nov 19. Am J Cardiol. 2016. PMID: 26708639
-
Rare genomic copy number variants implicate new candidate genes for bicuspid aortic valve.PLoS One. 2024 Sep 6;19(9):e0304514. doi: 10.1371/journal.pone.0304514. eCollection 2024. PLoS One. 2024. PMID: 39240962 Free PMC article.
-
Genetic Bases of Bicuspid Aortic Valve: The Contribution of Traditional and High-Throughput Sequencing Approaches on Research and Diagnosis.Front Physiol. 2017 Aug 24;8:612. doi: 10.3389/fphys.2017.00612. eCollection 2017. Front Physiol. 2017. PMID: 28883797 Free PMC article. Review.
-
The importance of genetics and genetic counselors in the evaluation of patients with bicuspid aortic valve and aortopathy.Curr Opin Cardiol. 2019 Jan;34(1):73-78. doi: 10.1097/HCO.0000000000000586. Curr Opin Cardiol. 2019. PMID: 30394908 Review.
Cited by
-
Risk estimation and risk prediction using machine-learning methods.Hum Genet. 2012 Oct;131(10):1639-54. doi: 10.1007/s00439-012-1194-y. Epub 2012 Jul 3. Hum Genet. 2012. PMID: 22752090 Free PMC article. Review.
-
Pathophysiological implications of different bicuspid aortic valve configurations.Cardiol Res Pract. 2012;2012:735829. doi: 10.1155/2012/735829. Epub 2012 May 27. Cardiol Res Pract. 2012. PMID: 22685682 Free PMC article.
-
Pathogenic Mechanisms of Bicuspid Aortic Valve Aortopathy.Front Physiol. 2017 Sep 25;8:687. doi: 10.3389/fphys.2017.00687. eCollection 2017. Front Physiol. 2017. PMID: 28993736 Free PMC article. Review.
-
Enlightening the Association between Bicuspid Aortic Valve and Aortopathy.J Cardiovasc Dev Dis. 2018 Apr 19;5(2):21. doi: 10.3390/jcdd5020021. J Cardiovasc Dev Dis. 2018. PMID: 29671812 Free PMC article. Review.
-
Identification of two novel mutations of the HOMEZ gene in Chinese patients with isolated ventricular septal defect.Genet Test Mol Biomarkers. 2013 May;17(5):390-4. doi: 10.1089/gtmb.2012.0435. Epub 2013 Apr 10. Genet Test Mol Biomarkers. 2013. PMID: 23574532 Free PMC article.
References
-
- Fedak PW, Verma S, David TE, Leask RL, Weisel RD, et al. Clinical and Pathophysiological Implications of a Bicuspid Aortic Valve. Circulation. 2002;106:900–904. - PubMed
-
- American Heart Association. Heart Disease and Stroke Statistics-2009 Update. 2009. Available at: http://www.americanheart.org/presenter.jhtml?identifier=3037327.
-
- Turri M, Thiene G, Bortolotti U, Milano A, Mazzucco A, et al. Surgical pathology of aortic valve disease. A study based on 602 specimens. Eur J Cardiothorac Surg. 1990;4:556–560. - PubMed
-
- Cripe L, Andelfinger G, Martin LJ, Shooner K, Benson DW. Bicuspid aortic valve is heritable. J Am Coll Cardiol. 2004;44:138–43. - PubMed
-
- Loscalzo ML, Goh DLM, Loeys B, Kent KC, Spevak PJ, et al. Familial thoracic aortic dilation and bicommissural aortic valve: a prospective analysis of natural history and inheritance. Am J Med Genet A. 2007;143:1960–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

