Parallel and convergent evolution of the dim-light vision gene RH1 in bats (Order: Chiroptera)
- PMID: 20098620
- PMCID: PMC2809114
- DOI: 10.1371/journal.pone.0008838
Parallel and convergent evolution of the dim-light vision gene RH1 in bats (Order: Chiroptera)
Abstract
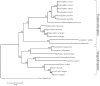
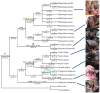
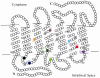
Rhodopsin, encoded by the gene Rhodopsin (RH1), is extremely sensitive to light, and is responsible for dim-light vision. Bats are nocturnal mammals that inhabit poor light environments. Megabats (Old-World fruit bats) generally have well-developed eyes, while microbats (insectivorous bats) have developed echolocation and in general their eyes were degraded, however, dramatic differences in the eyes, and their reliance on vision, exist in this group. In this study, we examined the rod opsin gene (RH1), and compared its evolution to that of two cone opsin genes (SWS1 and M/LWS). While phylogenetic reconstruction with the cone opsin genes SWS1 and M/LWS generated a species tree in accord with expectations, the RH1 gene tree united Pteropodidae (Old-World fruit bats) and Yangochiroptera, with very high bootstrap values, suggesting the possibility of convergent evolution. The hypothesis of convergent evolution was further supported when nonsynonymous sites or amino acid sequences were used to construct phylogenies. Reconstructed RH1 sequences at internal nodes of the bat species phylogeny showed that: (1) Old-World fruit bats share an amino acid change (S270G) with the tomb bat; (2) Miniopterus share two amino acid changes (V104I, M183L) with Rhinolophoidea; (3) the amino acid replacement I123V occurred independently on four branches, and the replacements L99M, L266V and I286V occurred each on two branches. The multiple parallel amino acid replacements that occurred in the evolution of bat RH1 suggest the possibility of multiple convergences of their ecological specialization (i.e., various photic environments) during adaptation for the nocturnal lifestyle, and suggest that further attention is needed on the study of the ecology and behavior of bats.
Conflict of interest statement
Figures




Similar articles
-
Molecular evolution of bat color vision genes.Mol Biol Evol. 2004 Feb;21(2):295-302. doi: 10.1093/molbev/msh015. Epub 2003 Dec 5. Mol Biol Evol. 2004. PMID: 14660703
-
The role of ecological factors in shaping bat cone opsin evolution.Proc Biol Sci. 2018 Apr 11;285(1876):20172835. doi: 10.1098/rspb.2017.2835. Proc Biol Sci. 2018. PMID: 29618549 Free PMC article.
-
Divergence of dim-light vision among bats (order: Chiroptera) as estimated by molecular and electrophysiological methods.Sci Rep. 2015 Jun 23;5:11531. doi: 10.1038/srep11531. Sci Rep. 2015. PMID: 26100095 Free PMC article.
-
The evolution of echolocation in bats.Trends Ecol Evol. 2006 Mar;21(3):149-56. doi: 10.1016/j.tree.2006.01.001. Epub 2006 Feb 8. Trends Ecol Evol. 2006. PMID: 16701491 Review.
-
Bat echolocation calls: adaptation and convergent evolution.Proc Biol Sci. 2007 Apr 7;274(1612):905-12. doi: 10.1098/rspb.2006.0200. Proc Biol Sci. 2007. PMID: 17251105 Free PMC article. Review.
Cited by
-
Molecular convergent and parallel evolution among four high-elevation anuran species from the Tibetan region.BMC Genomics. 2020 Nov 27;21(1):839. doi: 10.1186/s12864-020-07269-4. BMC Genomics. 2020. PMID: 33246413 Free PMC article.
-
The glycogen synthase 2 gene (Gys2) displays parallel evolution between Old World and New World fruit bats.J Mol Evol. 2014 Jan;78(1):66-74. doi: 10.1007/s00239-013-9600-1. Epub 2013 Nov 21. J Mol Evol. 2014. PMID: 24258790
-
Variation in opsin genes correlates with signalling ecology in North American fireflies.Mol Ecol. 2015 Sep;24(18):4679-96. doi: 10.1111/mec.13346. Mol Ecol. 2015. PMID: 26289828 Free PMC article.
-
Phosphoenolpyruvate carboxykinase 1 gene (Pck1) displays parallel evolution between Old World and New World fruit bats.PLoS One. 2015 Mar 25;10(3):e0118666. doi: 10.1371/journal.pone.0118666. eCollection 2015. PLoS One. 2015. PMID: 25807515 Free PMC article.
-
Variability in avian eggshell colour: a comparative study of museum eggshells.PLoS One. 2010 Aug 9;5(8):e12054. doi: 10.1371/journal.pone.0012054. PLoS One. 2010. PMID: 20711258 Free PMC article.
References
-
- Yokoyama S. Evolution of dim-light and color vision pigments. Annu Rev Genomics Hum Genet. 2008;9:259–282. - PubMed
-
- Yau KW. Phototransduction mechanism in retinal rods and cones. The Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1994;35:9–32. - PubMed
-
- Yokoyama S, Yokoyama R. Adaptive evolution of photoreceptors and visual pigments in vertebrates. Annu Rev Ecol Syst. 1996;27:543–567.
-
- Surridge AK, Osorio D, Mundy NI. Evolution and selection of trichromatic vision in primates. Trends Ecol Evol. 2003;18:198–205.
-
- Dominy NJ, Lucas PW. Ecological importance of trichromatic vision to primates. Nature. 2001;410:363–366. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

