ChIP-chip designs to interrogate the genome of Xenopus embryos for transcription factor binding and epigenetic regulation
- PMID: 20098671
- PMCID: PMC2809088
- DOI: 10.1371/journal.pone.0008820
ChIP-chip designs to interrogate the genome of Xenopus embryos for transcription factor binding and epigenetic regulation
Abstract
Background: Chromatin immunoprecipitation combined with genome tile path microarrays or deep sequencing can be used to study genome-wide epigenetic profiles and the transcription factor binding repertoire. Although well studied in a variety of cell lines, these genome-wide profiles have so far been little explored in vertebrate embryos.
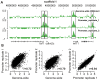
Principal findings: Here we report on two genome tile path ChIP-chip designs for interrogating the Xenopus tropicalis genome. In particular, a whole-genome microarray design was used to identify active promoters by close proximity to histone H3 lysine 4 trimethylation. A second microarray design features these experimentally derived promoter regions in addition to currently annotated 5' ends of genes. These regions truly represent promoters as shown by binding of TBP, a key transcription initiation factor.
Conclusions: A whole-genome and a promoter tile path microarray design was developed. Both designs can be used to study epigenetic phenomena and transcription factor binding in developing Xenopus embryos.
Conflict of interest statement
Figures

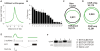
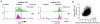
References
-
- Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. - PubMed
-
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. - PubMed
-
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. - PubMed
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

