Crystal structure and substrate specificity of Drosophila 3,4-dihydroxyphenylalanine decarboxylase
- PMID: 20098687
- PMCID: PMC2809104
- DOI: 10.1371/journal.pone.0008826
Crystal structure and substrate specificity of Drosophila 3,4-dihydroxyphenylalanine decarboxylase
Abstract
Background: 3,4-Dihydroxyphenylalanine decarboxylase (DDC), also known as aromatic L-amino acid decarboxylase, catalyzes the decarboxylation of a number of aromatic L-amino acids. Physiologically, DDC is responsible for the production of dopamine and serotonin through the decarboxylation of 3,4-dihydroxyphenylalanine and 5-hydroxytryptophan, respectively. In insects, both dopamine and serotonin serve as classical neurotransmitters, neuromodulators, or neurohormones, and dopamine is also involved in insect cuticle formation, eggshell hardening, and immune responses.
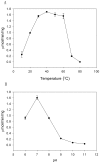
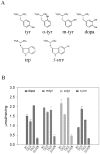
Principal findings: In this study, we expressed a typical DDC enzyme from Drosophila melanogaster, critically analyzed its substrate specificity and biochemical properties, determined its crystal structure at 1.75 Angstrom resolution, and evaluated the roles residues T82 and H192 play in substrate binding and enzyme catalysis through site-directed mutagenesis of the enzyme. Our results establish that this DDC functions exclusively on the production of dopamine and serotonin, with no activity to tyrosine or tryptophan and catalyzes the formation of serotonin more efficiently than dopamine.
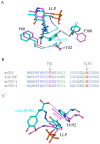
Conclusions: The crystal structure of Drosophila DDC and the site-directed mutagenesis study of the enzyme demonstrate that T82 is involved in substrate binding and that H192 is used not only for substrate interaction, but for cofactor binding of drDDC as well. Through comparative analysis, the results also provide insight into the structure-function relationship of other insect DDC-like proteins.
Conflict of interest statement
Figures
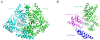
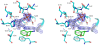
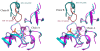
Similar articles
-
Biochemical identification of residues that discriminate between 3,4-dihydroxyphenylalanine decarboxylase and 3,4-dihydroxyphenylacetaldehyde synthase-mediated reactions.Insect Biochem Mol Biol. 2017 Dec;91:34-43. doi: 10.1016/j.ibmb.2017.10.001. Epub 2017 Oct 14. Insect Biochem Mol Biol. 2017. PMID: 29037755
-
Unraveling the Molecular Determinants of Catalytic Efficiency and Substrate Specificity in l-Amino Acid Decarboxylases.J Agric Food Chem. 2024 Dec 4;72(48):26996-27006. doi: 10.1021/acs.jafc.4c08560. Epub 2024 Nov 20. J Agric Food Chem. 2024. PMID: 39567248
-
Genomic organization of the rat aromatic L-amino acid decarboxylase (AADC) locus: partial analysis reveals divergence from the Drosophila dopa decarboxylase (DDC) gene structure.Mamm Genome. 1991;1(3):145-51. doi: 10.1007/BF00351060. Mamm Genome. 1991. PMID: 1797228
-
Aromatic Amino Acid Decarboxylase Deficiency: The Added Value of Biochemistry.Int J Mol Sci. 2021 Mar 19;22(6):3146. doi: 10.3390/ijms22063146. Int J Mol Sci. 2021. PMID: 33808712 Free PMC article. Review.
-
Plant aromatic L-amino acid decarboxylases: evolution, biochemistry, regulation, and metabolic engineering applications.Phytochemistry. 2000 May;54(2):121-38. doi: 10.1016/s0031-9422(00)00050-9. Phytochemistry. 2000. PMID: 10872203 Review.
Cited by
-
Construction of cell factory capable of efficiently converting L-tryptophan into 5-hydroxytryptamine.Microb Cell Fact. 2022 Mar 24;21(1):47. doi: 10.1186/s12934-022-01745-0. Microb Cell Fact. 2022. PMID: 35331215 Free PMC article.
-
Crystal structure of Oryza sativa TDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis.J Adv Res. 2020 Jun 12;24:501-511. doi: 10.1016/j.jare.2020.06.004. eCollection 2020 Jul. J Adv Res. 2020. PMID: 32595985 Free PMC article.
-
Identification and characterization of two arylalkylamine N-acetyltransferases in the yellow fever mosquito, Aedes aegypti.Insect Biochem Mol Biol. 2011 Sep;41(9):707-14. doi: 10.1016/j.ibmb.2011.05.002. Epub 2011 May 27. Insect Biochem Mol Biol. 2011. PMID: 21645618 Free PMC article.
-
Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.Proc Natl Acad Sci U S A. 2020 May 19;117(20):10806-10817. doi: 10.1073/pnas.1920097117. Epub 2020 May 5. Proc Natl Acad Sci U S A. 2020. PMID: 32371491 Free PMC article.
-
Mechanism-based tuning of insect 3,4-dihydroxyphenylacetaldehyde synthase for synthetic bioproduction of benzylisoquinoline alkaloids.Nat Commun. 2019 May 1;10(1):2015. doi: 10.1038/s41467-019-09610-2. Nat Commun. 2019. PMID: 31043610 Free PMC article.
References
-
- Sourkes TL. Dopa decarboxylase: substrates, coenzyme, inhibitors. Pharmacol Rev. 1966;18:53–60. - PubMed
-
- Hodgetts RB, O'Keefe SL. Dopa decarboxylase: a model gene-enzyme system for studying development, behavior, and systematics. Annu Rev Entomol. 2006;51:259–284. - PubMed
-
- Neckameyer WS, Leal SM. Biogenic amines as circulating hormones in insects. In: Pfaff DW, Arnold AP, Etgen AM, Fahrbach SE, Rubin RT, editors. Hormones, Brain and Behavior. San Diego: Academic; 2002. pp. 141–165.
-
- Nassel DR. Neuropeptides, amines and amino acids in an elementary insect ganglion: functional and chemical anatomy of the unfused abdominal ganglion. Prog Neurobiol. 1996;48:325–420. - PubMed
-
- Osborne RH. Insect neurotransmission: Neurotransmitters and their receptors. Pharmacology & Therapeutics. 1996;69:117–142. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

