The site specific demethylation in the 5'-regulatory area of NMDA receptor 2B subunit gene associated with CIE-induced up-regulation of transcription
- PMID: 20098704
- PMCID: PMC2808353
- DOI: 10.1371/journal.pone.0008798
The site specific demethylation in the 5'-regulatory area of NMDA receptor 2B subunit gene associated with CIE-induced up-regulation of transcription
Abstract
Background: The NMDA receptor represents a particularly important site of ethanol action in the CNS. We recently reported that NMDA receptor 2B (NR2B) gene expression was persistently up-regulated following chronic intermittent ethanol (CIE) treatment. Increasing evidence that epigenetic mechanisms are involved in dynamic and long-lasting regulation of gene expression in multiple neuroadaptive processes prompted us to investigate the role of DNA methylation in mediating CIE-induced up-regulation of NR2B gene transcription. To dissect the changes of DNA methylation in the NR2B gene, we have screened a large number of CpG sites within its 5'-regulatory area following CIE treatment.
Methods: Primary cortical cultured neurons were subjected to ethanol treatment in a CIE paradigm. Bisulfite conversion followed by pyrosequencing was used for quantitative measurement and analysis of CpG methylation status within the 5'-regulatory area of the NR2B gene; chromatin immunoprecipitation (ChIP) assay was used to examine DNA levels associated with methylation and transcription factor binding. Electrophoretic mobility shift assay (EMSA) and in vitro DNA methylation assays were performed to determine the direct impact of DNA methylation on the interaction between DNA and transcription factor and promoter activity.
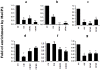
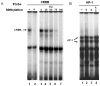
Results: Analysis of individual CpG methylation sites within the NR2B 5'regulatory area revealed three regions with clusters of site-specific CpG demethylation following CIE treatment and withdrawal. This was confirmed by ChIP showing similar decreases of methylated DNA in the same regions. The CIE-induced demethylation is characterized by being located near certain transcription factor binding sequences, AP-1 and CRE, and occurred during treatment as well as after ethanol withdrawal. Furthermore, the increase in vitro of methylated DNA decreased transcription factor binding activity and promoter activity. An additional ChIP assay indicated that the CIE-induced DNA demethylation is accompanied by increased occupation by transcription factors.
Conclusions: These results suggest an important role of DNA demethylation in mediating CIE-induced NR2B gene up-regulation, thus implicating a novel molecular site of alcohol action.
Conflict of interest statement
Figures

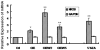

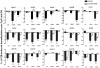
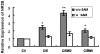
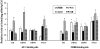
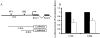

References
-
- Cagetti E, Liang J, Spigelman I, Olsen RW. Withdrawal from chronic intermittent ethanol treatment changes subunit composition, reduces synaptic function, and decreases behavioral responses to positive allosteric modulators of GABAA receptors. Mol Pharmacol. 2003;63:53–64. - PubMed
-
- Kokka N, Sapp DW, Taylor AM, Olsen RW. The kindling model of alcohol dependence: similar persistent reduction in seizure threshold to pentylenetetrazol in animals receiving chronic ethanol or chronic pentylenetetrazol. Alcohol Clin Exp Res. 1993;17:525–531. - PubMed
-
- Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci. 2001;2:119–128. - PubMed
-
- Ron D, Jurd R. The “ups and downs” of signaling cascades in addiction. Sci STKE 2005: re14. 2005:re14. - PubMed
-
- Nieuwenhuys R. The neocortex. An overview of its evolutionary development, structural organization and synaptology. Anat Embryol (Berl) 1994;190:307–337. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

