Genomic profiling of messenger RNAs and microRNAs reveals potential mechanisms of TWEAK-induced skeletal muscle wasting in mice
- PMID: 20098732
- PMCID: PMC2808241
- DOI: 10.1371/journal.pone.0008760
Genomic profiling of messenger RNAs and microRNAs reveals potential mechanisms of TWEAK-induced skeletal muscle wasting in mice
Abstract
Background: Skeletal muscle wasting is a devastating complication of several physiological and pathophysiological conditions. Inflammatory cytokines play an important role in the loss of skeletal muscle mass in various chronic diseases. We have recently reported that proinflammatory cytokine TWEAK is a major muscle-wasting cytokine. Emerging evidence suggests that gene expression is regulated not only at transcriptional level but also at post-transcriptional level through the expression of specific non-coding microRNAs (miRs) which can affect the stability and/or translation of target mRNA. However, the role of miRs in skeletal muscle wasting is unknown.
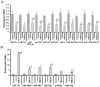
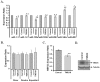
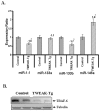
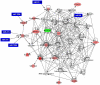
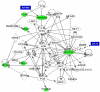
Methodology/principal findings: To understand the mechanism of action of TWEAK in skeletal muscle, we performed mRNA and miRs expression profile of control and TWEAK-treated myotubes. TWEAK increased the expression of a number of genes involved in inflammatory response and fibrosis and reduced the expression of few cytoskeletal gene (e.g. Myh4, Ankrd2, and TCap) and metabolic enzymes (e.g. Pgam2). Low density miR array demonstrated that TWEAK inhibits the expression of several miRs including muscle-specific miR-1-1, miR-1-2, miR-133a, miR-133b and miR-206. The expression of a few miRs including miR-146a and miR-455 was found to be significantly increased in response to TWEAK treatment. Ingenuity pathway analysis showed that several genes affected by TWEAK are known/putative targets of miRs. Our cDNA microarray data are consistent with miRs profiling. The levels of specific mRNAs and miRs were also found to be similarly regulated in atrophying skeletal muscle of transgenic mice (Tg) mice expressing TWEAK.
Conclusions/significance: Our results suggest that TWEAK affects the expression of several genes and microRNAs involved in inflammatory response, fibrosis, extracellular matrix remodeling, and proteolytic degradation which might be responsible for TWEAK-induced skeletal muscle loss.
Conflict of interest statement
Figures
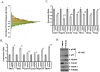

Similar articles
-
Genetic ablation of TWEAK augments regeneration and post-injury growth of skeletal muscle in mice.Am J Pathol. 2010 Oct;177(4):1732-42. doi: 10.2353/ajpath.2010.100335. Epub 2010 Aug 19. Am J Pathol. 2010. PMID: 20724600 Free PMC article.
-
Elevated levels of TWEAK in skeletal muscle promote visceral obesity, insulin resistance, and metabolic dysfunction.FASEB J. 2015 Mar;29(3):988-1002. doi: 10.1096/fj.14-260703. Epub 2014 Dec 2. FASEB J. 2015. PMID: 25466899 Free PMC article.
-
The TWEAK-Fn14 system: breaking the silence of cytokine-induced skeletal muscle wasting.Curr Mol Med. 2012 Jan;12(1):3-13. doi: 10.2174/156652412798376107. Curr Mol Med. 2012. PMID: 22082477 Free PMC article. Review.
-
The TWEAK-Fn14 system is a critical regulator of denervation-induced skeletal muscle atrophy in mice.J Cell Biol. 2010 Mar 22;188(6):833-49. doi: 10.1083/jcb.200909117. J Cell Biol. 2010. PMID: 20308426 Free PMC article.
-
MitomiRs in human inflamm-aging: a hypothesis involving miR-181a, miR-34a and miR-146a.Exp Gerontol. 2014 Aug;56:154-63. doi: 10.1016/j.exger.2014.03.002. Epub 2014 Mar 7. Exp Gerontol. 2014. PMID: 24607549 Review.
Cited by
-
Gene profiling studies in skeletal muscle by quantitative real-time polymerase chain reaction assay.Methods Mol Biol. 2012;798:311-24. doi: 10.1007/978-1-61779-343-1_18. Methods Mol Biol. 2012. PMID: 22130845 Free PMC article.
-
Skeletal Muscle Remodelling as a Function of Disease Progression in Amyotrophic Lateral Sclerosis.Biomed Res Int. 2016;2016:5930621. doi: 10.1155/2016/5930621. Epub 2016 Apr 18. Biomed Res Int. 2016. PMID: 27195289 Free PMC article.
-
TWEAK/Fn14 Signaling Axis Mediates Skeletal Muscle Atrophy and Metabolic Dysfunction.Front Immunol. 2014 Jan 27;5:18. doi: 10.3389/fimmu.2014.00018. eCollection 2014. Front Immunol. 2014. PMID: 24478779 Free PMC article. Review.
-
Integrative Analysis of MicroRNA and mRNA Data Reveals an Orchestrated Function of MicroRNAs in Skeletal Myocyte Differentiation in Response to TNF-α or IGF1.PLoS One. 2015 Aug 13;10(8):e0135284. doi: 10.1371/journal.pone.0135284. eCollection 2015. PLoS One. 2015. PMID: 26270642 Free PMC article.
-
miR-133b Inhibits Cell Growth, Migration, and Invasion by Targeting MMP9 in Non-Small Cell Lung Cancer.Oncol Res. 2017 Aug 7;25(7):1109-1116. doi: 10.3727/096504016X14800889609439. Epub 2016 Nov 29. Oncol Res. 2017. PMID: 27938481 Free PMC article.
References
-
- Acharyya S, Guttridge DC. Cancer cachexia signaling pathways continue to emerge yet much still points to the proteasome. Clin Cancer Res. 2007;13:1356–1361. - PubMed
-
- Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol. 2004;287:C834–843. - PubMed
-
- Spate U, Schulze PC. Proinflammatory cytokines and skeletal muscle. Curr Opin Clin Nutr Metab Care. 2004;7:265–269. - PubMed
-
- Tracey KJ, Cerami A. Tumor necrosis factor, other cytokines and disease. Annu Rev Cell Biol. 1993;9:317–343. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

