Concordant association of insulin degrading enzyme gene (IDE) variants with IDE mRNA, Abeta, and Alzheimer's disease
- PMID: 20098734
- PMCID: PMC2808243
- DOI: 10.1371/journal.pone.0008764
Concordant association of insulin degrading enzyme gene (IDE) variants with IDE mRNA, Abeta, and Alzheimer's disease
Abstract
Background: The insulin-degrading enzyme gene (IDE) is a strong functional and positional candidate for late onset Alzheimer's disease (LOAD).
Methodology/principal findings: We examined conserved regions of IDE and its 10 kb flanks in 269 AD cases and 252 controls thereby identifying 17 putative functional polymorphisms. These variants formed eleven haplotypes that were tagged with ten variants. Four of these showed significant association with IDE transcript levels in samples from 194 LOAD cerebella. The strongest, rs6583817, which has not previously been reported, showed unequivocal association (p = 1.5x10(-8), fold-increase = 2.12,); the eleven haplotypes were also significantly associated with transcript levels (global p = 0.003). Using an in vitro dual luciferase reporter assay, we found that rs6583817 increases reporter gene expression in Be(2)-C (p = 0.006) and HepG2 (p = 0.02) cell lines. Furthermore, using data from a recent genome-wide association study of two Croatian isolated populations (n = 1,879), we identified a proxy for rs6583817 that associated significantly with decreased plasma Abeta40 levels (ss = -0.124, p = 0.011) and total measured plasma Abeta levels (b = -0.130, p = 0.009). Finally, rs6583817 was associated with decreased risk of LOAD in 3,891 AD cases and 3,605 controls. (OR = 0.87, p = 0.03), and the eleven IDE haplotypes (global p = 0.02) also showed significant association.
Conclusions: Thus, a previously unreported variant unequivocally associated with increased IDE expression was also associated with reduced plasma Abeta40 and decreased LOAD susceptibility. Genetic association between LOAD and IDE has been difficult to replicate. Our findings suggest that targeted testing of expression SNPs (eSNPs) strongly associated with altered transcript levels in autopsy brain samples may be a powerful way to identify genetic associations with LOAD that would otherwise be difficult to detect.
Conflict of interest statement
Figures

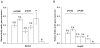

References
-
- Walsh DM, Selkoe DJ. Deciphering the molecular basis of memory failure in Alzheimer's disease. Neuron. 2004;44:181–193. - PubMed
-
- Bertram L, Tanzi RE. The current status of Alzheimer's disease genetics: what do we tell the patients? Pharmacol Res. 2004;50:385–396. - PubMed
-
- Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, et al. Heritability for Alzheimer's disease: the study of dementia in Swedish twins. J Gerontol A Biol Sci Med Sci. 1997;52:M117–125. - PubMed
-
- Bergem AL, Engedal K, Kringlen E. The role of heredity in late-onset Alzheimer disease and vascular dementia. A twin study. Arch Gen Psychiatry. 1997;54:264–270. - PubMed
-
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 AG16574/AG/NIA NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- MC_U127561128/MRC_/Medical Research Council/United Kingdom
- KL2 RR024151/RR/NCRR NIH HHS/United States
- R01 AG032990/AG/NIA NIH HHS/United States
- P01 AG003949/AG/NIA NIH HHS/United States
- P01 AG017216/AG/NIA NIH HHS/United States
- R01 AG018023/AG/NIA NIH HHS/United States
- AG17216/AG/NIA NIH HHS/United States
- AG03949/AG/NIA NIH HHS/United States
- F32 AG020903/AG/NIA NIH HHS/United States
- U01 AG06576/AG/NIA NIH HHS/United States
- AG25711/AG/NIA NIH HHS/United States
- R01 AG18023/AG/NIA NIH HHS/United States
- F32 AG20903/AG/NIA NIH HHS/United States
- U24 AG21886/AG/NIA NIH HHS/United States
- P50 AG025711/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

