3'-end sequencing for expression quantification (3SEQ) from archival tumor samples
- PMID: 20098735
- PMCID: PMC2808244
- DOI: 10.1371/journal.pone.0008768
3'-end sequencing for expression quantification (3SEQ) from archival tumor samples
Abstract
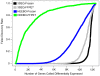
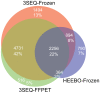
Gene expression microarrays are the most widely used technique for genome-wide expression profiling. However, microarrays do not perform well on formalin fixed paraffin embedded tissue (FFPET). Consequently, microarrays cannot be effectively utilized to perform gene expression profiling on the vast majority of archival tumor samples. To address this limitation of gene expression microarrays, we designed a novel procedure (3'-end sequencing for expression quantification (3SEQ)) for gene expression profiling from FFPET using next-generation sequencing. We performed gene expression profiling by 3SEQ and microarray on both frozen tissue and FFPET from two soft tissue tumors (desmoid type fibromatosis (DTF) and solitary fibrous tumor (SFT)) (total n = 23 samples, which were each profiled by at least one of the four platform-tissue preparation combinations). Analysis of 3SEQ data revealed many genes differentially expressed between the tumor types (FDR<0.01) on both the frozen tissue (approximately 9.6K genes) and FFPET (approximately 8.1K genes). Analysis of microarray data from frozen tissue revealed fewer differentially expressed genes (approximately 4.64K), and analysis of microarray data on FFPET revealed very few (69) differentially expressed genes. Functional gene set analysis of 3SEQ data from both frozen tissue and FFPET identified biological pathways known to be important in DTF and SFT pathogenesis and suggested several additional candidate oncogenic pathways in these tumors. These findings demonstrate that 3SEQ is an effective technique for gene expression profiling from archival tumor samples and may facilitate significant advances in translational cancer research.
Conflict of interest statement
Figures



References
-
- Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. - PubMed
-
- Lipshutz RJ, Fodor SPA, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nature genetics. 1999;21:20–24. - PubMed
-
- Potti A, Dressman HK, Bild A, Riedel RF, Chan G, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. - PubMed
-
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. - PubMed
-
- Van't Veer LJ, Dai H, Van de Vijver MJ, He YD, Hart AA, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 415:530. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

