Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships
- PMID: 20100353
- PMCID: PMC2823734
- DOI: 10.1186/1471-2148-10-26
Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships
Abstract
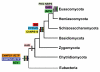
Background: Nonribosomal peptide synthetases (NRPSs) are multimodular enzymes, found in fungi and bacteria, which biosynthesize peptides without the aid of ribosomes. Although their metabolite products have been the subject of intense investigation due to their life-saving roles as medicinals and injurious roles as mycotoxins and virulence factors, little is known of the phylogenetic relationships of the corresponding NRPSs or whether they can be ranked into subgroups of common function. We identified genes (NPS) encoding NRPS and NRPS-like proteins in 38 fungal genomes and undertook phylogenomic analyses in order to identify fungal NRPS subfamilies, assess taxonomic distribution, evaluate levels of conservation across subfamilies, and address mechanisms of evolution of multimodular NRPSs. We also characterized relationships of fungal NRPSs, a representative sampling of bacterial NRPSs, and related adenylating enzymes, including alpha-aminoadipate reductases (AARs) involved in lysine biosynthesis in fungi.
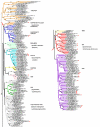
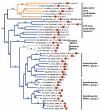
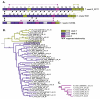
Results: Phylogenomic analysis identified nine major subfamilies of fungal NRPSs which fell into two main groups: one corresponds to NPS genes encoding primarily mono/bi-modular enzymes which grouped with bacterial NRPSs and the other includes genes encoding primarily multimodular and exclusively fungal NRPSs. AARs shared a closer phylogenetic relationship to NRPSs than to other acyl-adenylating enzymes. Phylogenetic analyses and taxonomic distribution suggest that several mono/bi-modular subfamilies arose either prior to, or early in, the evolution of fungi, while two multimodular groups appear restricted to and expanded in fungi. The older mono/bi-modular subfamilies show conserved domain architectures suggestive of functional conservation, while multimodular NRPSs, particularly those unique to euascomycetes, show a diversity of architectures and of genetic mechanisms generating this diversity.
Conclusions: This work is the first to characterize subfamilies of fungal NRPSs. Our analyses suggest that mono/bi-modular NRPSs have more ancient origins and more conserved domain architectures than most multimodular NRPSs. It also demonstrates that the alpha-aminoadipate reductases involved in lysine biosynthesis in fungi are closely related to mono/bi-modular NRPSs. Several groups of mono/bi-modular NRPS metabolites are predicted to play more pivotal roles in cellular metabolism than products of multimodular NRPSs. In contrast, multimodular subfamilies of NRPSs are of more recent origin, are restricted to fungi, show less stable domain architectures, and biosynthesize metabolites which perform more niche-specific functions than mono/bi-modular NRPS products. The euascomycete-only NRPS subfamily, in particular, shows evidence for extensive gain and loss of domains suggestive of the contribution of domain duplication and loss in responding to niche-specific pressures.
Figures






Similar articles
-
Module evolution and substrate specificity of fungal nonribosomal peptide synthetases involved in siderophore biosynthesis.BMC Evol Biol. 2008 Dec 3;8:328. doi: 10.1186/1471-2148-8-328. BMC Evol Biol. 2008. PMID: 19055762 Free PMC article.
-
Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins.Toxins (Basel). 2013 Apr 19;5(4):717-42. doi: 10.3390/toxins5040717. Toxins (Basel). 2013. PMID: 23604065 Free PMC article.
-
Bacterial-Like Nonribosomal Peptide Synthetases Produce Cyclopeptides in the Zygomycetous Fungus Mortierella alpina.Appl Environ Microbiol. 2021 Jan 15;87(3):e02051-20. doi: 10.1128/AEM.02051-20. Print 2021 Jan 15. Appl Environ Microbiol. 2021. PMID: 33158886 Free PMC article.
-
Nonribosomal Peptide Synthetases in Animals.Genes (Basel). 2023 Aug 30;14(9):1741. doi: 10.3390/genes14091741. Genes (Basel). 2023. PMID: 37761881 Free PMC article. Review.
-
New insights into the echinocandins and other fungal non-ribosomal peptides and peptaibiotics.Nat Prod Rep. 2014 Oct;31(10):1348-75. doi: 10.1039/c4np00046c. Nat Prod Rep. 2014. PMID: 25156669 Review.
Cited by
-
Genetic engineering activates biosynthesis of aromatic fumaric acid amides in the human pathogen Aspergillus fumigatus.Appl Environ Microbiol. 2015 Mar;81(5):1594-600. doi: 10.1128/AEM.03268-14. Epub 2014 Dec 19. Appl Environ Microbiol. 2015. PMID: 25527545 Free PMC article.
-
Multifunctional polyketide synthase genes identified by genomic survey of the symbiotic dinoflagellate, Symbiodinium minutum.BMC Genomics. 2015 Nov 14;16:941. doi: 10.1186/s12864-015-2195-8. BMC Genomics. 2015. PMID: 26573520 Free PMC article.
-
Quorum sensing regulators and non-ribosomal peptide synthetases govern antibacterial secretions in Xenorhabdus szentirmaii.Front Microbiol. 2025 Mar 12;16:1560663. doi: 10.3389/fmicb.2025.1560663. eCollection 2025. Front Microbiol. 2025. PMID: 40143860 Free PMC article.
-
Individual and combined roles of malonichrome, ferricrocin, and TAFC siderophores in Fusarium graminearum pathogenic and sexual development.Front Microbiol. 2015 Jan 12;5:759. doi: 10.3389/fmicb.2014.00759. eCollection 2014. Front Microbiol. 2015. PMID: 25628608 Free PMC article.
-
Next-generation sequencing approach for connecting secondary metabolites to biosynthetic gene clusters in fungi.Front Microbiol. 2015 Jan 14;5:774. doi: 10.3389/fmicb.2014.00774. eCollection 2014. Front Microbiol. 2015. PMID: 25642215 Free PMC article. Review.
References
-
- Stein T, Vater J, Kruft V, Otto A, Wittmann-Liebold B, Franke P, Panico M, McDowell R, Morris HR. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. Journal of Biological Chemistry. 1996;271(26):15428–15435. doi: 10.1074/jbc.271.26.15428. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

