Constitutively overexpressed 21 kDa protein in Hodgkin lymphoma and aggressive non-Hodgkin lymphomas identified as cytochrome B5b (CYB5B)
- PMID: 20100355
- PMCID: PMC2829491
- DOI: 10.1186/1476-4598-9-14
Constitutively overexpressed 21 kDa protein in Hodgkin lymphoma and aggressive non-Hodgkin lymphomas identified as cytochrome B5b (CYB5B)
Abstract
Background: We have previously reported a novel constitutively overexpressed 21 kDa protein in Hodgkin Lymphoma (HL) and aggressive Non-Hodgkin Lymphomas (NHL). The objective of the current study was to 1) identify this protein using two independent methods, 2) study the expression of the protein and its encoding mRNA in reactive lymph nodes, normal lymphocytes and CD34+ bone marrow precursor cells, 3) analyse patterns of expression of the protein in tissue microarrays assembled from a large number of diagnostic clinical biopsies from patients with HL, and 4) determine the copy number variation and mutation status of the encoding gene in HL cell lines.
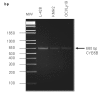
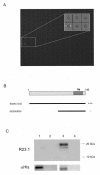
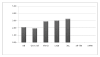
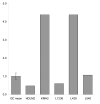
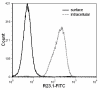
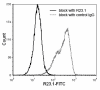
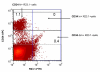
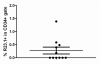
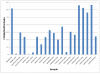
Results: Peptide sequencing by LC-MS/MS and protein identification by protein array screening identified a single protein, CYB5B. No mutations were detected in the CYB5B gene in HL cell lines. Quantitative PCR showed CYB5B gene expression was increased in HL and NHL cell lines. Array CGH using a submegabase resolution tiling array revealed gains in the CYB5B locus in HL cell lines KMH2 and L428. Membrane expression was seen in Reed-Sternberg cells in clinical biopsies from patients with HL but not in reactive lymph nodes. Bone marrow CD34+ precursor cells were CYB5B negative on the cell surface. RT-PCR assays of RNA extracted from T and B cell enriched fractions obtained from normal peripheral blood mononuclear cells, reactive lymph nodes, tonsils and normal bone marrow samples showed no evidence of increased mRNA levels of CYB5B in comparison to housekeeping gene GAPDH.
Conclusions: The 21 kDa protein overexpressed in HL and aggressive NHL is identical to CYB5B. CYB5B gene expression is increased in a subset of HL and NHL cell lines tested. This is associated with CYB5B gene amplification in HL cell lines KMH2 and L428. CYB5B may be a potential target for antibody-based therapy of HL and aggressive NHL as although cytoplasmic expression is present in reactive lymphocytes, it is not expressed on the cell surface of non-neoplastic lymphocytes or bone marrow precursor cells.
Figures








Similar articles
-
Constitutive overexpression of a novel 21 kDa protein by Hodgkin lymphoma and aggressive non-Hodgkin lymphomas.Mol Cancer. 2008 Jan 24;7:12. doi: 10.1186/1476-4598-7-12. Mol Cancer. 2008. PMID: 18218123 Free PMC article.
-
Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma.Blood. 2002 Jan 15;99(2):618-26. doi: 10.1182/blood.v99.2.618. Blood. 2002. PMID: 11781246
-
Sub-megabase resolution tiling (SMRT) array-based comparative genomic hybridization profiling reveals novel gains and losses of chromosomal regions in Hodgkin Lymphoma and Anaplastic Large Cell Lymphoma cell lines.Mol Cancer. 2008 Jan 7;7:2. doi: 10.1186/1476-4598-7-2. Mol Cancer. 2008. PMID: 18179710 Free PMC article.
-
Hodgkin and non-Hodgkin lymphoma of the head and neck: clinical, pathologic, and imaging evaluation.Neuroimaging Clin N Am. 2003 Aug;13(3):371-92. doi: 10.1016/s1052-5149(03)00039-x. Neuroimaging Clin N Am. 2003. PMID: 14631680 Review.
-
Reed-Sternberg-Like Cells in Non-Hodgkin Lymphomas.Arch Pathol Lab Med. 2015 Oct;139(10):1205-10. doi: 10.5858/arpa.2015-0197-RAI. Arch Pathol Lab Med. 2015. PMID: 26414463 Review.
Cited by
-
Consensus reference gene(s) for gene expression studies in human cancers: end of the tunnel visible?Cell Oncol (Dordr). 2015 Dec;38(6):419-31. doi: 10.1007/s13402-015-0244-6. Epub 2015 Sep 18. Cell Oncol (Dordr). 2015. PMID: 26384826 Review.
-
Anti-proliferative effect and underlying mechanism of ethoxy-substituted phylloquinone (vitamin K1 derivative) from Spinacia oleracea leaf and enhancement of its extractability using radiation technology.3 Biotech. 2022 Oct;12(10):265. doi: 10.1007/s13205-022-03264-6. Epub 2022 Sep 8. 3 Biotech. 2022. PMID: 36091087 Free PMC article.
-
Simultaneous structure-activity studies and arming of natural products by C-H amination reveal cellular targets of eupalmerin acetate.Nat Chem. 2013 Jun;5(6):510-7. doi: 10.1038/nchem.1653. Epub 2013 May 19. Nat Chem. 2013. PMID: 23695633 Free PMC article.
-
Metformin Therapeutic Targets for Aortic Aneurysms: A Mendelian Randomization and Colocalization Study.Rev Cardiovasc Med. 2024 Mar 5;25(3):89. doi: 10.31083/j.rcm2503089. eCollection 2024 Mar. Rev Cardiovasc Med. 2024. PMID: 39076954 Free PMC article.
-
Discovery of Potent and Selective Inhibitors against Protein-Derived Electrophilic Cofactors.J Am Chem Soc. 2022 Mar 30;144(12):5377-5388. doi: 10.1021/jacs.1c12748. Epub 2022 Mar 2. J Am Chem Soc. 2022. PMID: 35235319 Free PMC article.
References
-
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumours. Vol. 2. IARC; 2008.
-
- Jundt F, Acikgoz O, Kwon SH, Schwarzer R, Anagnostopoulos I, Wiesner B, Mathas S, Hummel M, Stein H, Reichardt HM, Dorken B. Aberrant expression of Notch1 interferes with the B-lymphoid phenotype of neoplastic B cells in classical Hodgkin lymphoma. Leukemia. 2008;22:1587–1594. doi: 10.1038/leu.2008.101. - DOI - PubMed
-
- Doerr JR, Malone CS, Fike FM, Gordon MS, Soghomonian SV, Thomas RK, Tao Q, Murray PG, Diehl V, Teitell MA, Wall R. Patterned CpG methylation of silenced B cell gene promoters in classical Hodgkin lymphoma-derived and primary effusion lymphoma cell lines. J Mol Biol. 2005;350:631–640. doi: 10.1016/j.jmb.2005.05.032. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

