Cooperativity within proximal phosphorylation sites is revealed from large-scale proteomics data
- PMID: 20100358
- PMCID: PMC2828979
- DOI: 10.1186/1745-6150-5-6
Cooperativity within proximal phosphorylation sites is revealed from large-scale proteomics data
Abstract
Background: Phosphorylation is the most prevalent post-translational modification on eukaryotic proteins. Multisite phosphorylation enables a specific combination of phosphosites to determine the speed, specificity and duration of biological response. Until recent years, the lack of high quality data limited the possibility for analyzing the properties of phosphorylation at the proteome scale and in the context of a wide range of conditions. Thanks to advances of mass spectrometry technologies, thousands of phosphosites from in-vivo experiments were identified and archived in the public domain. Such resource is appropriate to derive an unbiased view on the phosphosites properties in eukaryotes and on their functional relevance.
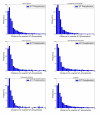
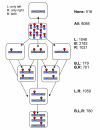
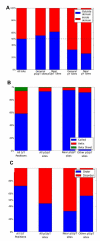
Results: We present statistically rigorous tests on the spatial and functional properties of a collection of approximately 70,000 reported phosphosites. We show that the distribution of phosphosites positioning along the protein tends to occur as dense clusters of Serine/Threonines (pS/pT) and between Serine/Threonines and Tyrosines, but generally not as much between Tyrosines (pY) only. This phenomenon is more ubiquitous than anticipated and is pertinent for most eukaryotic proteins: for proteins with > or = 2 phosphosites, 54% of all pS/pT sites are within 4 amino acids of another site. We found a strong tendency for clustered pS/pT to be activated by the same kinase. Large-scale analyses of phosphopeptides are thus consistent with a cooperative function within the cluster.
Conclusions: We present evidence supporting the notion that clusters of pS/pT but generally not pY should be considered as the elementary building blocks in phosphorylation regulation. Indeed, closely positioned sites tend to be activated by the same kinase, a signal that overrides the tendency of a protein to be activated by a single or only few kinases. Within these clusters, coordination and positional dependency is evident. We postulate that cellular regulation takes advantage of such design. Specifically, phosphosite clusters may increase the robustness of the effectiveness of phosphorylation-dependent response.
Reviewers: Reviewed by Joel Bader, Frank Eisenhaber, Emmanuel Levy (nominated by Sarah Teichmann). For the full reviews, please go to the Reviewers' comments section.
Figures



References
-
- Turkina MV, Vener AV. Identification of phosphorylated proteins. Methods Mol Biol. 2007;355:305–316. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

