Expression profiling and comparative analyses of seven midgut serine proteases from the yellow fever mosquito, Aedes aegypti
- PMID: 20100490
- PMCID: PMC2878907
- DOI: 10.1016/j.jinsphys.2010.01.003
Expression profiling and comparative analyses of seven midgut serine proteases from the yellow fever mosquito, Aedes aegypti
Abstract
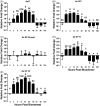
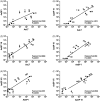
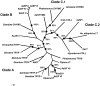
Aedes aegypti utilizes blood for energy production, egg maturation and replenishment of maternal reserves. The principle midgut enzymes responsible for bloodmeal digestion are endoproteolytic serine-type proteases within the S1.A subfamily. While there are hundreds of serine protease-like genes in the A. aegypti genome, only five are known to be expressed in the midgut. We describe the cloning, sequencing and expression profiling of seven additional serine proteases and provide a genomic and phylogenetic assessment of these findings. Of the seven genes, four are constitutively expressed and three are transcriptionally induced upon blood feeding. The amount of transcriptional induction is strongly correlated among these genes. Alignments reveal that, in general, the conserved catalytic triad, active site and accessory catalytic residues are maintained in these genes and phylogenetic analysis shows that these genes fall within three distinct clades; trypsins, chymotrypsins and serine collagenases. Interestingly, a previously described trypsin consistently arose with other serine collagenases in phylogenetic analyses. These results suggest that multiple gene duplications have arisen within the S1.A subfamily of midgut serine proteases and/or that A. aegypti has evolved an array of proteases with a broad range of substrate specificities for rapid, efficient digestion of bloodmeals.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
In vitro activation and enzyme kinetic analysis of recombinant midgut serine proteases from the Dengue vector mosquito Aedes aegypti.BMC Biochem. 2011 Aug 9;12:43. doi: 10.1186/1471-2091-12-43. BMC Biochem. 2011. PMID: 21827688 Free PMC article.
-
Molecular genetic analysis of midgut serine proteases in Aedes aegypti mosquitoes.Insect Biochem Mol Biol. 2009 Dec;39(12):903-12. doi: 10.1016/j.ibmb.2009.10.008. Epub 2009 Nov 3. Insect Biochem Mol Biol. 2009. PMID: 19883761 Free PMC article.
-
E93 confers steroid hormone responsiveness of digestive enzymes to promote blood meal digestion in the midgut of the mosquito Aedes aegypti.Insect Biochem Mol Biol. 2021 Jul;134:103580. doi: 10.1016/j.ibmb.2021.103580. Epub 2021 Apr 24. Insect Biochem Mol Biol. 2021. PMID: 33901693 Free PMC article.
-
Molecular analysis of the Aedes aegypti carboxypeptidase gene family.Insect Biochem Mol Biol. 2009 Jan;39(1):68-73. doi: 10.1016/j.ibmb.2008.09.006. Epub 2008 Oct 14. Insect Biochem Mol Biol. 2009. PMID: 18977440 Free PMC article.
-
Isolation, sequencing and characterization of two cDNA clones coding for trypsin-like enzymes from the midgut of Aedes aegypti.Insect Mol Biol. 1993;2(2):71-9. doi: 10.1111/j.1365-2583.1993.tb00127.x. Insect Mol Biol. 1993. PMID: 9087545
Cited by
-
Transcriptomic analyses of Aedes aegypti cultured cells and ex vivo midguts in response to an excess or deficiency of heme: a quest for transcriptionally-regulated heme transporters.BMC Genomics. 2020 Aug 31;21(1):604. doi: 10.1186/s12864-020-06981-5. BMC Genomics. 2020. PMID: 32867680 Free PMC article.
-
Exome and transcriptome sequencing of Aedes aegypti identifies a locus that confers resistance to Brugia malayi and alters the immune response.PLoS Pathog. 2015 Mar 27;11(3):e1004765. doi: 10.1371/journal.ppat.1004765. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25815506 Free PMC article.
-
Determination of juvenile hormone titers by means of LC-MS/MS/MS and a juvenile hormone-responsive Gal4/UAS system in Aedes aegypti mosquitoes.Insect Biochem Mol Biol. 2016 Oct;77:69-77. doi: 10.1016/j.ibmb.2016.08.003. Epub 2016 Aug 12. Insect Biochem Mol Biol. 2016. PMID: 27530057 Free PMC article.
-
In-depth characterization of trypsin-like serine peptidases in the midgut of the sugar fed Culex quinquefasciatus.Parasit Vectors. 2015 Jul 16;8:373. doi: 10.1186/s13071-015-0985-0. Parasit Vectors. 2015. PMID: 26174750 Free PMC article.
-
A transcriptomic atlas of Aedes aegypti reveals detailed functional organization of major body parts and gut regional specializations in sugar-fed and blood-fed adult females.Elife. 2022 Apr 26;11:e76132. doi: 10.7554/eLife.76132. Elife. 2022. PMID: 35471187 Free PMC article.
References
-
- Barillas-Mury C, Graf R, Hagedorn HH, Wells MA. cDNA and deduced amino acid sequence of a blood meal-induced trypsin from the mosquito Aedes aegypti. Insect Biochemistry. 1991;21:825–831.
-
- Barillas-Mury C, Wells MA. Cloning and sequencing of the blood meal-induced late trypsin gene from the mosquito Aedes aegypti and characterization of the upstream regulatory region. Insect Molecular Biology. 1993;2:7–12. - PubMed
-
- Barros C, Crosby JA, Moreno RD. Early steps of sperm–egg interactions during mammalian fertilization. Cell Biology International. 1996;20:33–39. - PubMed
-
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340:783–795. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

