Outer membrane protein I of Pseudomonas aeruginosa is a target of cationic antimicrobial peptide/protein
- PMID: 20100832
- PMCID: PMC2838320
- DOI: 10.1074/jbc.M109.078725
Outer membrane protein I of Pseudomonas aeruginosa is a target of cationic antimicrobial peptide/protein
Abstract
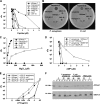
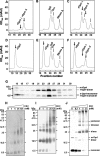
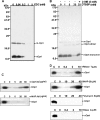
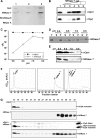
Cationic antimicrobial peptides/proteins (AMPs) are important components of the host innate defense mechanisms against invading microorganisms. Here we demonstrate that OprI (outer membrane protein I) of Pseudomonas aeruginosa is responsible for its susceptibility to human ribonuclease 7 (hRNase 7) and alpha-helical cationic AMPs, instead of surface lipopolysaccharide, which is the initial binding site of cationic AMPs. The antimicrobial activities of hRNase 7 and alpha-helical cationic AMPs against P. aeruginosa were inhibited by the addition of exogenous OprI or anti-OprI antibody. On modification and internalization of OprI by hRNase 7 into cytosol, the bacterial membrane became permeable to metabolites. The lipoprotein was predicted to consist of an extended loop at the N terminus for hRNase 7/lipopolysaccharide binding, a trimeric alpha-helix, and a lysine residue at the C terminus for cell wall anchoring. Our findings highlight a novel mechanism of antimicrobial activity and document a previously unexplored target of alpha-helical cationic AMPs, which may be used for screening drugs to treat antibiotic-resistant bacterial infection.
Figures




Similar articles
-
Key Residues of Outer Membrane Protein OprI Involved in Hexamer Formation and Bacterial Susceptibility to Cationic Antimicrobial Peptides.Antimicrob Agents Chemother. 2015 Oct;59(10):6210-22. doi: 10.1128/AAC.01406-15. Epub 2015 Jul 27. Antimicrob Agents Chemother. 2015. PMID: 26248382 Free PMC article.
-
Hydrophobic residues are critical for the helix-forming, hemolytic and bactericidal activities of amphipathic antimicrobial peptide TP4.PLoS One. 2017 Oct 17;12(10):e0186442. doi: 10.1371/journal.pone.0186442. eCollection 2017. PLoS One. 2017. PMID: 29040295 Free PMC article.
-
Sarkosyl-Induced Helical Structure of an Antimicrobial Peptide GW-Q6 Plays an Essential Role in the Binding of Surface Receptor OprI in Pseudomonas aeruginosa.PLoS One. 2016 Oct 11;11(10):e0164597. doi: 10.1371/journal.pone.0164597. eCollection 2016. PLoS One. 2016. PMID: 27727309 Free PMC article.
-
NMR Structures and Interactions of Antimicrobial Peptides with Lipopolysaccharide: Connecting Structures to Functions.Curr Top Med Chem. 2016;16(1):4-15. doi: 10.2174/1568026615666150703121943. Curr Top Med Chem. 2016. PMID: 26139110 Review.
-
An intimate link between antimicrobial peptide sequence diversity and binding to essential components of bacterial membranes.Biochim Biophys Acta. 2016 May;1858(5):958-70. doi: 10.1016/j.bbamem.2015.10.011. Epub 2015 Oct 21. Biochim Biophys Acta. 2016. PMID: 26498397 Review.
Cited by
-
Efficient photodynamic therapy against gram-positive and gram-negative bacteria using THPTS, a cationic photosensitizer excited by infrared wavelength.PLoS One. 2010 Jul 20;5(7):e11674. doi: 10.1371/journal.pone.0011674. PLoS One. 2010. PMID: 20652031 Free PMC article.
-
Potent bactericidal activity of reduced cryptdin-4 derived from its hydrophobicity and mediated by bacterial membrane disruption.Amino Acids. 2022 Feb;54(2):289-297. doi: 10.1007/s00726-021-03115-3. Epub 2022 Jan 17. Amino Acids. 2022. PMID: 35037097
-
A bovine myeloid antimicrobial peptide (BMAP-28) and its analogs kill pan-drug-resistant Acinetobacter baumannii by interacting with outer membrane protein A (OmpA).Medicine (Baltimore). 2018 Oct;97(42):e12832. doi: 10.1097/MD.0000000000012832. Medicine (Baltimore). 2018. PMID: 30334982 Free PMC article.
-
Molecular Detection of Drug-Resistance Genes of blaOXA-23-blaOXA-51 and mcr-1 in Clinical Isolates of Pseudomonas aeruginosa.Microorganisms. 2021 Apr 9;9(4):786. doi: 10.3390/microorganisms9040786. Microorganisms. 2021. PMID: 33918745 Free PMC article.
-
Max Bergmann lecture protein epitope mimetics in the age of structural vaccinology.J Pept Sci. 2013 Mar;19(3):127-40. doi: 10.1002/psc.2482. Epub 2013 Jan 24. J Pept Sci. 2013. PMID: 23349031 Free PMC article. Review.
References
-
- Giamarellou H. (2002) J. Antimicrob. Chemother. 49, 229–233 - PubMed
-
- Zasloff M. (2002) Nature 415, 389–395 - PubMed
-
- Brown K. L., Hancock R. E. (2006) Curr. Opin. Immunol. 18, 24–30 - PubMed
-
- Bowdish D. M., Davidson D. J., Hancock R. E. (2005) Curr. Protein Pept. Sci. 6, 35–51 - PubMed
-
- Gläser R., Harder J., Lange H., Bartels J., Christophers E., Schröder J. M. (2005) Nat. Immunol. 6, 57–64 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

