DNA breaks at fragile sites generate oncogenic RET/PTC rearrangements in human thyroid cells
- PMID: 20101222
- PMCID: PMC2855398
- DOI: 10.1038/onc.2009.502
DNA breaks at fragile sites generate oncogenic RET/PTC rearrangements in human thyroid cells
Abstract
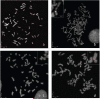
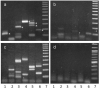
Human chromosomal fragile sites are regions of the genome that are prone to DNA breakage, and are classified as common or rare, depending on their frequency in the population. Common fragile sites frequently coincide with the location of genes involved in carcinogenic chromosomal translocations, suggesting their role in cancer formation. However, there has been no direct evidence linking breakage at fragile sites to the formation of a cancer-specific translocation. Here, we studied the involvement of fragile sites in the formation of RET/PTC rearrangements, which are frequently found in papillary thyroid carcinoma (PTC). These rearrangements are commonly associated with radiation exposure; however, most of the tumors found in adults are not linked to radiation. In this study, we provide structural and biochemical evidence that the RET, CCDC6 and NCOA4 genes participating in two major types of RET/PTC rearrangements, are located in common fragile sites FRA10C and FRA10G, and undergo DNA breakage after exposure to fragile site-inducing chemicals. Moreover, exposure of human thyroid cells to these chemicals results in the formation of cancer-specific RET/PTC rearrangements. These results provide the direct evidence for the involvement of chromosomal fragile sites in the generation of cancer-specific rearrangements in human cells.
Figures


Similar articles
-
DNA fragile site breakage as a measure of chemical exposure and predictor of individual susceptibility to form oncogenic rearrangements.Carcinogenesis. 2017 Mar 1;38(3):293-301. doi: 10.1093/carcin/bgw210. Carcinogenesis. 2017. PMID: 28069693 Free PMC article.
-
DNA topoisomerases participate in fragility of the oncogene RET.PLoS One. 2013 Sep 11;8(9):e75741. doi: 10.1371/journal.pone.0075741. eCollection 2013. PLoS One. 2013. PMID: 24040417 Free PMC article.
-
A post-irradiation-induced replication stress promotes RET proto-oncogene breakage.Eur Thyroid J. 2024 Aug 23;13(4):e240028. doi: 10.1530/ETJ-24-0028. Print 2024 Aug 1. Eur Thyroid J. 2024. PMID: 39047147 Free PMC article.
-
[RET/PTC Gene Rearrangements in the Sporadic and Radiogenic Thyroid Tumors: Molecular Genetics, Radiobiology and Molecular Epidemiology].Radiats Biol Radioecol. 2015 May-Jun;55(3):229-49. Radiats Biol Radioecol. 2015. PMID: 26310016 Review. Russian.
-
Mechanisms of chromosomal rearrangements in solid tumors: the model of papillary thyroid carcinoma.Mol Cell Endocrinol. 2010 May 28;321(1):36-43. doi: 10.1016/j.mce.2009.09.013. Epub 2009 Sep 18. Mol Cell Endocrinol. 2010. PMID: 19766698 Free PMC article. Review.
Cited by
-
Extended RET gene analysis in patients with apparently sporadic medullary thyroid cancer: clinical benefits and cost.Horm Cancer. 2012 Aug;3(4):181-6. doi: 10.1007/s12672-012-0109-7. Epub 2012 May 31. Horm Cancer. 2012. PMID: 22648435 Free PMC article.
-
The role of fragile sites in sporadic papillary thyroid carcinoma.J Thyroid Res. 2012;2012:927683. doi: 10.1155/2012/927683. Epub 2012 Jun 11. J Thyroid Res. 2012. PMID: 22762011 Free PMC article.
-
Triggers for genomic rearrangements: insights into genomic, cellular and environmental influences.Nat Rev Genet. 2010 Dec;11(12):819-29. doi: 10.1038/nrg2883. Epub 2010 Nov 3. Nat Rev Genet. 2010. PMID: 21045868 Review.
-
RET Gene Fusions in Malignancies of the Thyroid and Other Tissues.Genes (Basel). 2020 Apr 15;11(4):424. doi: 10.3390/genes11040424. Genes (Basel). 2020. PMID: 32326537 Free PMC article. Review.
-
Role of DNA secondary structures in fragile site breakage along human chromosome 10.Hum Mol Genet. 2013 Apr 1;22(7):1443-56. doi: 10.1093/hmg/dds561. Epub 2013 Jan 7. Hum Mol Genet. 2013. PMID: 23297364 Free PMC article.
References
-
- Arlt MF, Durkin SG, Ragland RL, Glover TW. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst) 2006;5:1126–35. - PubMed
-
- Boldog F, Gemmill RM, West J, Robinson M, Robinson L, Li E, et al. Chromosome 3p14 homozygous deletions and sequence analysis of FRA3B. Hum Mol Genet. 1997;6:193–203. - PubMed
-
- Bongarzone I, Butti MG, Fugazzola L, Pacini F, Pinchera A, Vorontsova TV, et al. Comparison of the breakpoint regions of ELE1 and RET genes involved in the generation of RET/PTC3 oncogene in sporadic and in radiation-associated papillary thyroid carcinomas. Genomics. 1997;42:252–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

