A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33
- PMID: 20101243
- PMCID: PMC2853179
- DOI: 10.1038/ng.522
A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33
Abstract
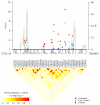
We conducted a genome-wide association study of pancreatic cancer in 3,851 affected individuals (cases) and 3,934 unaffected controls drawn from 12 prospective cohort studies and 8 case-control studies. Based on a logistic regression model for genotype trend effect that was adjusted for study, age, sex, self-described ancestry and five principal components, we identified eight SNPs that map to three loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Two correlated SNPs, rs9543325 (P = 3.27 x 10(-11), per-allele odds ratio (OR) 1.26, 95% CI 1.18-1.35) and rs9564966 (P = 5.86 x 10(-8), per-allele OR 1.21, 95% CI 1.13-1.30), map to a nongenic region on chromosome 13q22.1. Five SNPs on 1q32.1 map to NR5A2, and the strongest signal was at rs3790844 (P = 2.45 x 10(-10), per-allele OR 0.77, 95% CI 0.71-0.84). A single SNP, rs401681 (P = 3.66 x 10(-7), per-allele OR 1.19, 95% CI 1.11-1.27), maps to the CLPTM1L-TERT locus on 5p15.33, which is associated with multiple cancers. Our study has identified common susceptibility loci for pancreatic cancer that warrant follow-up studies.
Figures


References
-
- Ferlay J, Bray F, Pisani P, Parkin DM. IARC CancerBase. No. 5. IARCPress; Lyon: 2004. GLOBOCAN 2002: Cancer Incidence, Mortality and Prevalence Worldwide.
-
- Anderson KE,MT, Silverman D. Cancer of the pancreas. In: Schottenfeld D, Fraumeni JJ, editors. Cancer Epidemiology and Prevention. Oxford University Press; New York: 2006.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

