Systems analysis of EGF receptor signaling dynamics with microwestern arrays
- PMID: 20101245
- PMCID: PMC2881471
- DOI: 10.1038/nmeth.1418
Systems analysis of EGF receptor signaling dynamics with microwestern arrays
Abstract
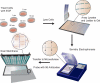
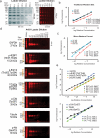
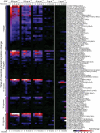
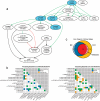
We describe microwestern arrays, which enable quantitative, sensitive and high-throughput assessment of protein abundance and modifications after electrophoretic separation of microarrayed cell lysates. This method allowed us to measure 91 phosphosites on 67 proteins at six time points after stimulation with five epidermal growth factor (EGF) concentrations in A431 human carcinoma cells. We inferred the connectivities among 15 phosphorylation sites in 10 receptor tyrosine kinases (RTKs) and two sites from Src kinase using Bayesian network modeling and two mutual information-based methods; the three inference methods yielded substantial agreement on the network topology. These results imply multiple distinct RTK coactivation mechanisms and support the notion that small amounts of experimental data collected from phenotypically diverse network states may enable network inference.
Figures


References
-
- Burnette W. Western Blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Analytical Biochemistry. 1981;112:195–203. - PubMed
-
- Paweletz CP, Liotta LA, Petricoin EF. New technologies for biomarker analysis of prostate cancer progression: Laser capture microdissection and tissue proteomics. Urology. 2001;57:160–3. - PubMed
-
- Paweletz CP, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20:1981–9. - PubMed
-
- Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal Protein-Signaling Networks Derived from Multiparameter Single-Cell Data. Science. 2005;308:523–529. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

