A guided tour of the Trans-Proteomic Pipeline
- PMID: 20101611
- PMCID: PMC3017125
- DOI: 10.1002/pmic.200900375
A guided tour of the Trans-Proteomic Pipeline
Abstract
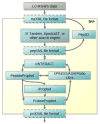
The Trans-Proteomic Pipeline (TPP) is a suite of software tools for the analysis of MS/MS data sets. The tools encompass most of the steps in a proteomic data analysis workflow in a single, integrated software system. Specifically, the TPP supports all steps from spectrometer output file conversion to protein-level statistical validation, including quantification by stable isotope ratios. We describe here the full workflow of the TPP and the tools therein, along with an example on a sample data set, demonstrating that the setup and use of the tools are straightforward and well supported and do not require specialized informatic resources or knowledge.
Figures

References
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
-
- Bantscheff M, et al. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389(4):1017–31. - PubMed
-
- Deutsch EW, Lam H, Aebersold R. Data analysis and bioinformatics tools for tandem mass spectrometry in proteomics. Physiol Genomics. 2008;33(1):18–25. - PubMed
-
- Kohlbacher O, et al. TOPP--the OpenMS proteomics pipeline. Bioinformatics. 2007;23(2):e191–7. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

