CCA addition to tRNA: implications for tRNA quality control
- PMID: 20101632
- PMCID: PMC2848691
- DOI: 10.1002/iub.301
CCA addition to tRNA: implications for tRNA quality control
Abstract
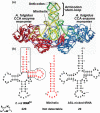
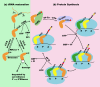
The CCA sequence is conserved at the 3' end of all mature tRNA molecules to function as the site of amino acid attachment. This sequence is acquired and maintained by stepwise nucleotide addition by the ubiquitous CCA enzyme, which is an unusual RNA polymerase that does not use a nucleic acid template for nucleotide addition. Crystal structural work has divided CCA enzymes into two structurally distinct classes, which differ in the mechanism of template-independent nucleotide selection. Recent kinetic work of the class II E. coli CCA enzyme has demonstrated a rapid and uniform rate constant for the chemistry of nucleotide addition at each step of CCA synthesis, although the enzyme uses different determinants to control the rate of each step. Importantly, the kinetic work reveals that, at each step of CCA synthesis, E. coli CCA enzyme has an innate ability to discriminate against tRNA backbone damage. This discrimination suggests the possibility of a previously unrecognized quality control mechanism that would prevent damaged tRNA from CCA maturation and from entering the ribosome machinery of protein synthesis. This quality control is relevant to cellular stress conditions that damage tRNA backbone and predicts a role of CCA addition in stress response.
Figures


Similar articles
-
tRNA integrity is a prerequisite for rapid CCA addition: implication for quality control.J Mol Biol. 2008 Jun 6;379(3):579-88. doi: 10.1016/j.jmb.2008.04.005. Epub 2008 Apr 8. J Mol Biol. 2008. PMID: 18466919 Free PMC article.
-
Distinct kinetic determinants for the stepwise CCA addition to tRNA.RNA. 2009 Oct;15(10):1827-36. doi: 10.1261/rna.1669109. Epub 2009 Aug 20. RNA. 2009. PMID: 19696158 Free PMC article.
-
Pyrophosphorolysis of CCA addition: implication for fidelity.J Mol Biol. 2011 Nov 18;414(1):28-43. doi: 10.1016/j.jmb.2011.09.036. Epub 2011 Oct 5. J Mol Biol. 2011. PMID: 22001019
-
The CCA-adding enzyme: A central scrutinizer in tRNA quality control.Bioessays. 2015 Sep;37(9):975-82. doi: 10.1002/bies.201500043. Epub 2015 Jul 14. Bioessays. 2015. PMID: 26172425 Review.
-
A story with a good ending: tRNA 3'-end maturation by CCA-adding enzymes.Curr Opin Struct Biol. 2006 Feb;16(1):12-7. doi: 10.1016/j.sbi.2005.12.001. Epub 2005 Dec 20. Curr Opin Struct Biol. 2006. PMID: 16364630 Review.
Cited by
-
Analysis of tRNA Cys processing under salt stress in Bacillus subtilis spore outgrowth using RNA sequencing data.F1000Res. 2020 Jun 3;9:501. doi: 10.12688/f1000research.23780.1. eCollection 2020. F1000Res. 2020. PMID: 33976872 Free PMC article.
-
Critical Minireview: The Fate of tRNACys during Oxidative Stress in Bacillus subtilis.Biomolecules. 2017 Jan 20;7(1):6. doi: 10.3390/biom7010006. Biomolecules. 2017. PMID: 28117687 Free PMC article. Review.
-
MINTmap: fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data.Sci Rep. 2017 Feb 21;7:41184. doi: 10.1038/srep41184. Sci Rep. 2017. PMID: 28220888 Free PMC article.
-
tRNA 3'-amino-tailing for stable amino acid attachment.RNA. 2018 Dec;24(12):1878-1885. doi: 10.1261/rna.068015.118. Epub 2018 Sep 14. RNA. 2018. PMID: 30217865 Free PMC article.
-
A Temporal Order in 5'- and 3'- Processing of Eukaryotic tRNAHis.Int J Mol Sci. 2019 Mar 19;20(6):1384. doi: 10.3390/ijms20061384. Int J Mol Sci. 2019. PMID: 30893886 Free PMC article.
References
-
- Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. - PubMed
-
- Tomari Y, Hino N, Nagaike T, Suzuki T, Ueda T. Decreased CCA-addition in human mitochondrial tRNAs bearing a pathogenic A4317G or A10044G mutation. J Biol Chem. 2003;278:16828–16833. - PubMed
-
- Levinger L, Oestreich I, Florentz C, Morl M. A pathogenesis-associated mutation in human mitochondrial tRNALeu(UUR) leads to reduced 3'-end processing and CCA addition. J Mol Biol. 2004;337:535–544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

