Novel enzymatic function of DNA polymerase nu in translesion DNA synthesis past major groove DNA-peptide and DNA-DNA cross-links
- PMID: 20102227
- PMCID: PMC2838406
- DOI: 10.1021/tx900449u
Novel enzymatic function of DNA polymerase nu in translesion DNA synthesis past major groove DNA-peptide and DNA-DNA cross-links
Abstract
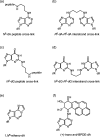
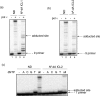
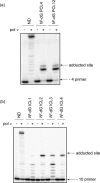
DNA polymerase nu (POLN or pol nu) is a newly discovered A family polymerase that generates a high error rate when incorporating nucleotides opposite dG; its translesion DNA synthesis (TLS) capability has only been demonstrated for high fidelity replication bypass of thymine glycol lesions. In the current investigation, we describe a novel TLS substrate specificity of pol nu, demonstrating that it is able to bypass exceptionally large DNA lesions whose linkages are through the DNA major groove. Specifically, pol nu catalyzed efficient and high fidelity TLS past peptides linked to N(6)-dA via a reduced Schiff base linkage with a gamma-hydroxypropano-dA. Additionally, pol nu could bypass DNA interstrand cross-links with linkage between N(6)-dAs in complementary DNA strands. However, the chemically identical DNA--peptide and DNA interstrand cross-links completely blocked pol nu when they were located in the minor groove via a N(2)-dG linkage. Furthermore, we showed that pol nu incorporated a nucleotide opposite the 1,N(6)-etheno-dA (epsilondA) in an error-free manner and (+)-trans-anti-benzo[a]pyrene-7,8-dihydrodiol 9,10-epoxide-dA [(+)-BPDE-dA] in an error-prone manner, albeit with a greatly reduced capability. Collectively, these data suggest that although pol nu bypass capacity cannot be generalized to all major groove DNA adducts, this polymerase could be involved in TLS when genomic replication is blocked by extremely large major groove DNA lesions. In view of the recent observation that pol nu may have a role in cellular tolerance to DNA cross-linking agents, our findings provide biochemical evidence for the potential functioning of this polymerase in the bypass of some DNA-protein and DNA-DNA cross-links.
Figures



Similar articles
-
Replication bypass of N2-deoxyguanosine interstrand cross-links by human DNA polymerases η and ι.Chem Res Toxicol. 2012 Mar 19;25(3):755-62. doi: 10.1021/tx300011w. Epub 2012 Feb 29. Chem Res Toxicol. 2012. PMID: 22332732 Free PMC article.
-
Mutagenic Bypass of an Oxidized Abasic Lesion-Induced DNA Interstrand Cross-Link Analogue by Human Translesion Synthesis DNA Polymerases.Biochemistry. 2015 Dec 22;54(50):7409-22. doi: 10.1021/acs.biochem.5b01027. Epub 2015 Dec 14. Biochemistry. 2015. PMID: 26626537 Free PMC article.
-
Role of high-fidelity Escherichia coli DNA polymerase I in replication bypass of a deoxyadenosine DNA-peptide cross-link.J Bacteriol. 2011 Aug;193(15):3815-21. doi: 10.1128/JB.01550-10. Epub 2011 May 27. J Bacteriol. 2011. PMID: 21622737 Free PMC article.
-
A Comprehensive View of Translesion Synthesis in Escherichia coli.Microbiol Mol Biol Rev. 2020 Jun 17;84(3):e00002-20. doi: 10.1128/MMBR.00002-20. Print 2020 Aug 19. Microbiol Mol Biol Rev. 2020. PMID: 32554755 Free PMC article. Review.
-
Translesion DNA synthesis polymerases in DNA interstrand crosslink repair.Environ Mol Mutagen. 2010 Jul;51(6):552-66. doi: 10.1002/em.20573. Environ Mol Mutagen. 2010. PMID: 20658647 Review.
Cited by
-
Synthesis and polymerase bypass studies of DNA-peptide and DNA-protein conjugates.Methods Enzymol. 2021;661:363-405. doi: 10.1016/bs.mie.2021.09.005. Epub 2021 Oct 26. Methods Enzymol. 2021. PMID: 34776221 Free PMC article.
-
DNA polymerases and cancer.Nat Rev Cancer. 2011 Feb;11(2):96-110. doi: 10.1038/nrc2998. Nat Rev Cancer. 2011. PMID: 21258395 Free PMC article. Review.
-
Involvement of translesion synthesis DNA polymerases in DNA interstrand crosslink repair.DNA Repair (Amst). 2016 Aug;44:33-41. doi: 10.1016/j.dnarep.2016.05.004. Epub 2016 May 13. DNA Repair (Amst). 2016. PMID: 27311543 Free PMC article. Review.
-
Synthesis of structurally diverse major groove DNA interstrand crosslinks using three different aldehyde precursors.Nucleic Acids Res. 2014 Jun;42(11):7429-35. doi: 10.1093/nar/gku328. Epub 2014 Apr 29. Nucleic Acids Res. 2014. PMID: 24782532 Free PMC article.
-
Analysis of DNA polymerase ν function in meiotic recombination, immunoglobulin class-switching, and DNA damage tolerance.PLoS Genet. 2017 Jun 1;13(6):e1006818. doi: 10.1371/journal.pgen.1006818. eCollection 2017 Jun. PLoS Genet. 2017. PMID: 28570559 Free PMC article.
References
-
- Marini F.; Kim N.; Schuffert A.; Wood R. D. (2003) POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 278, 32014–32019. - PubMed
-
- Takata K.; Shimizu T.; Iwai S.; Wood R. D. (2006) Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J. Biol. Chem. 281, 23445–23455. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

