Neuropilin-1 regulates platelet-derived growth factor receptor signalling in mesenchymal stem cells
- PMID: 20102335
- PMCID: PMC3441150
- DOI: 10.1042/BJ20091512
Neuropilin-1 regulates platelet-derived growth factor receptor signalling in mesenchymal stem cells
Abstract
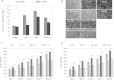
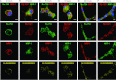
Using human MSCs (mesenchymal stem cells) lacking VEGF (vascular endothelial growth factor) receptors, we show that the pro-angiogenic receptor neuropilin-1 associates with phosphorylated PDGFRs [PDGF (platelet-derived growth factor) receptors], thereby regulating cell signalling, migration, proliferation and network assembly. Neuropilin-1 co-immunoprecipitated and co-localized with phosphorylated PDGFRs in the presence of growth factors. Neuropilin-1 knockdown blocked PDGF-AA-induced PDGFRalpha phosphorylation and migration, reduced PDGF-BB-induced PDGFRbeta activation and migration, blocked VEGF-A activation of both PDGFRs, and attenuated proliferation. Neuropilin-1 prominently co-localized with both PDGFRs within MSC networks assembled in Matrigel and in the chorioallantoic membrane vasculature microenvironment, and its knockdown grossly disrupted network assembly and decreased PDGFR signalling. Thus neuropilin-1 regulates MSCs by forming ligand-specific receptor complexes that direct PDGFR signalling, especially the PDGFRalpha homodimer. This receptor cross-talk may control the mobilization of MSCs in neovascularization and tissue remodelling.
Figures






References
-
- Oswald J., Boxberger S., Jorgensen B., Feldmann S., Ehninger G., Bornhauser M., Werner C. Mesenchymal stem cells can differentiate into endothelial cells in vitro. Stem Cells. 2004;22:377–384. - PubMed
-
- Chen M. Y., Lie P. C., Li Z. L., Wei X. Endothelial differentiation of Wharton's jelly-derived mesenchymal stem cells in comparison with bone marrow-derived mesenchymal stem cells. Exp. Hematol. 2009;37:629–640. - PubMed
-
- Ball S. G., Shuttleworth C. A., Kielty C. M. Platelet-derived growth factor receptor-α is a key determinant of smooth muscle α-actin in bone marrow-derived mesenchymal stem cells. Int. J. Biochem. Cell Biol. 2007;39:379–391.. - PubMed
-
- Chen H., Chedotal A., He Z., Goodman C. S., Tessier-Lavigne M. Neuropilin-2, a novel member of the neuropilin family, is a high affinity receptor for the semaphorins Sema E and Sema IV but not Sema III. Neuron. 1997;19:547–559. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

