Sex and the single embryo: early deveiopment in the Mediterranean fruit fly, Ceratitis capitata
- PMID: 20102629
- PMCID: PMC2826288
- DOI: 10.1186/1471-213X-10-12
Sex and the single embryo: early deveiopment in the Mediterranean fruit fly, Ceratitis capitata
Abstract
Background: In embryos the maternal-to-zygotic transition (MTZ) integrates post-transcriptional regulation of maternal transcripts with transcriptional activation of the zygotic genome. Although the molecular mechanisms underlying this event are being clarified in Drosophila melanogaster, little is know about the embryogenic processes in other insect species. The recent publication of expressed sequence tags (ESTs) from embryos of the global pest species Ceratitis capitata (medfly) has enabled the investigation of embryogenesis in this species and has allowed a comparison of the embryogenic processes in these two related dipteran species, C. capitata and D. melanogaster, that shared a common ancestor 80-100 mya.
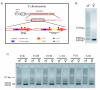
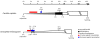
Results: Using a novel PCR-based sexing method, which takes advantage of a putative LTR retrotransposon MITE insertion on the medfly Y chromosome, the transcriptomes of individual early male and female embryos were analysed using RT-PCR. This study is focused on two crucial aspects of the onset of embryonic development: sex determination and cellular blastoderm formation. Together with the three known medfly genes (Cctransformer, Cctransformer2 and Ccdoublesex), the expression patterns of other medfly genes that are similar to the D. melanogaster sex-determination genes (sisterlessA, groucho, deadpan, Sex-lethal, female lethal d, sans fille and intersex) and four cellular blastoderm formation genes (Rho1, spaghetti squash, slow-as-molasses and serendipity-alpha) were analyzed, allowing us to sketch a preliminary outline of the embryonic process in the medfly. Furthermore, a putative homologue of the Zelda gene has been considered, which in D. melanogaster encodes a DNA-binding factor responsible for the maternal-to-zygotic transition.
Conclusions: Our novel sexing method facilitates the study of i) when the MTZ transition occurs in males and females of C. capitata, ii) when and how the maternal information of "female-development" is reprogrammed in the embryos and iii) similarities and differences in the regulation of gene expression in C. capitata and D. melanogaster. We suggest a new model for the onset of the sex determination cascade in the medfly: the maternally inherited Cctra transcripts in the female embryos are insufficient to produce enough active protein to inhibit the male mode of Cctra splicing. The slow rate of development and the inefficiency of the splicing mechanism in the pre-cellular blastoderm facilitates the male-determining factor (M) activity, which probably acts by inhibiting CcTRA protein activity.
Figures




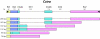
References
-
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell. 2007;12(1):143–155. doi: 10.1016/j.devcel.2006.10.005. - DOI - PubMed

