Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes
- PMID: 20103589
- PMCID: PMC2847748
- DOI: 10.1101/gr.098947.109
Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes
Abstract
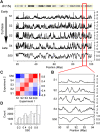
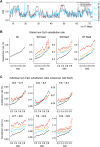
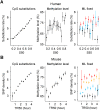
Neutral nucleotide substitutions occur at varying rates along genomes, and it remains a major issue to unravel the mechanisms that cause these variations and to analyze their evolutionary consequences. Here, we study the role of replication in the neutral substitution pattern. We obtained a high-resolution replication timing profile of the whole human genome by massively parallel sequencing of nascent BrdU-labeled replicating DNA. These data were compared to the neutral substitution rates along the human genome, obtained by aligning human and chimpanzee genomes using macaque and orangutan as outgroups. All substitution rates increase monotonously with replication timing even after controlling for local or regional nucleotide composition, crossover rate, distance to telomeres, and chromatin compaction. The increase in non-CpG substitution rates might result from several mechanisms including the increase in mutation-prone activities or the decrease in efficiency of DNA repair during the S phase. In contrast, the rate of C --> T transitions in CpG dinucleotides increases in later-replicating regions due to increasing DNA methylation level that reflects a negative correlation between timing and gene expression. Similar results are observed in the mouse, which indicates that replication timing is a main factor affecting nucleotide substitution dynamics at non-CpG sites and constitutes a major neutral process driving mammalian genome evolution.
Figures

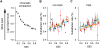
References
-
- Azuara V. Profiling of DNA replication timing in unsynchronized cell populations. Nat Protoc. 2006;1:2171–2177. - PubMed
-
- Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. - PubMed
-
- Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
