Acute inhibition of fatty acid import inhibits GLUT4 transcription in adipose tissue, but not skeletal or cardiac muscle tissue, partly through liver X receptor (LXR) signaling
- PMID: 20103707
- PMCID: PMC2844827
- DOI: 10.2337/db09-1542
Acute inhibition of fatty acid import inhibits GLUT4 transcription in adipose tissue, but not skeletal or cardiac muscle tissue, partly through liver X receptor (LXR) signaling
Abstract
Objective: Insulin-mediated glucose uptake is highly sensitive to the levels of the facilitative GLUT protein GLUT4. Transcription of the GLUT4 gene is repressed in states of insulin deficiency and insulin resistance and can be induced by states of enhanced energy output, such as exercise. The cellular signals that regulate GLUT4 transcription are not well understood. We hypothesized that changes in energy substrate flux regulate GLUT4 transcription.
Research design and methods: To test this hypothesis, we used transgenic mice in which expression of the chloramphenicol acetyltransferase (CAT) gene is driven by a functional 895-bp fragment of the human GLUT4 promoter, thereby acting as a reporter for transcriptional activity. Mice were treated with a single dose of etomoxir, which inhibits the transport of long-chain fatty acids into mitochondria and increases basal, but not insulin-mediated, glucose flux. GLUT4 and transgenic CAT mRNA were measured.
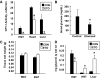
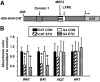
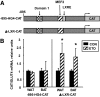
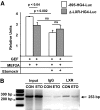
Results: Etomoxir treatment significantly reduced CAT and GLUT4 mRNA transcription in adipose tissue, but did not change transcription in heart and skeletal muscle. Downregulation of GLUT4 transcription was cell autonomous, since etomoxir treatment of 3T3-L1 adipocytes resulted in a similar downregulation of GLUT4 mRNA. GLUT4 transcriptional downregulation required the putative liver X receptor (LXR) binding site in the human GLUT4 gene promoter in adipose tissue and 3T3-L1 adipocytes. Treatment of 3T3-L1 adipocytes with the LXR agonist, TO901317, partially restored GLUT4 expression in etomoxir-treated cells.
Conclusions: Our data suggest that long-chain fatty acid import into mitochondria in adipose tissue may produce ligands that regulate expression of metabolic genes.
Figures



Similar articles
-
Expression of the insulin-responsive glucose transporter GLUT4 in adipocytes is dependent on liver X receptor alpha.J Biol Chem. 2003 Nov 28;278(48):48283-91. doi: 10.1074/jbc.M302287200. Epub 2003 Sep 11. J Biol Chem. 2003. PMID: 12970362
-
Expression and regulation of the human GLUT4/muscle-fat facilitative glucose transporter gene in transgenic mice.J Biol Chem. 1992 Jun 15;267(17):11673-6. J Biol Chem. 1992. PMID: 1601840
-
Hormonal/metabolic regulation of the human GLUT4/muscle-fat facilitative glucose transporter gene in transgenic mice.J Biol Chem. 1993 May 5;268(13):9839-46. J Biol Chem. 1993. PMID: 8486663
-
Class II histone deacetylases downregulate GLUT4 transcription in response to increased cAMP signaling in cultured adipocytes and fasting mice.Diabetes. 2012 Jun;61(6):1404-14. doi: 10.2337/db11-0737. Epub 2012 Mar 8. Diabetes. 2012. PMID: 22403301 Free PMC article.
-
Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue.Proc Natl Acad Sci U S A. 2003 Apr 29;100(9):5419-24. doi: 10.1073/pnas.0830671100. Epub 2003 Apr 15. Proc Natl Acad Sci U S A. 2003. PMID: 12697904 Free PMC article.
Cited by
-
Class II histone deacetylases limit GLUT4 gene expression during adipocyte differentiation.J Biol Chem. 2011 Jan 7;286(1):460-8. doi: 10.1074/jbc.M110.157107. Epub 2010 Nov 3. J Biol Chem. 2011. PMID: 21047791 Free PMC article.
-
LXRα improves myocardial glucose tolerance and reduces cardiac hypertrophy in a mouse model of obesity-induced type 2 diabetes.Diabetologia. 2016 Mar;59(3):634-43. doi: 10.1007/s00125-015-3827-x. Epub 2015 Dec 18. Diabetologia. 2016. PMID: 26684450 Free PMC article.
-
Glucose availability controls adipogenesis in mouse 3T3-L1 adipocytes via up-regulation of nicotinamide metabolism.J Biol Chem. 2017 Nov 10;292(45):18556-18564. doi: 10.1074/jbc.M117.791970. Epub 2017 Sep 15. J Biol Chem. 2017. PMID: 28916720 Free PMC article.
-
Increased cardiac PFK-2 protects against high-fat diet-induced cardiomyopathy and mediates beneficial systemic metabolic effects.iScience. 2023 Jun 15;26(7):107131. doi: 10.1016/j.isci.2023.107131. eCollection 2023 Jul 21. iScience. 2023. PMID: 37534142 Free PMC article.
-
Spatiotemporal dynamics of triglyceride storage in unilocular adipocytes.Mol Biol Cell. 2014 Dec 15;25(25):4096-105. doi: 10.1091/mbc.E14-06-1085. Epub 2014 Oct 8. Mol Biol Cell. 2014. PMID: 25298400 Free PMC article.
References
-
- Tsao TS, Burcelin R, Katz EB, Huang L, Charron MJ: Enhanced insulin action due to targeted GLUT4 overexpression exclusively in muscle. Diabetes 1996; 45: 28– 36 - PubMed
-
- Treadway JL, Hargrove DM, Nardone NA, McPherson RK, Russo JF, Milici AJ, Stukenbrok HA, Gibbs EM, Stevenson RW, Pessin JE: Enhanced peripheral glucose utilization in transgenic mice expressing the human GLUT4 gene. J Biol Chem 1994; 269: 29956– 29961 - PubMed
-
- Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, Kahn BB: Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J Biol Chem 1993; 268: 22243– 22246 - PubMed
-
- Katz EB, Stenbit AE, Hatton K, DePinho R, Charron MJ: Cardiac and adipose tissue abnormalities but not diabetes in mice deficient in GLUT4. Nature 1995; 377: 151– 155 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

