HIV sequence variation associated with env antisense adoptive T-cell therapy in the hNSG mouse model
- PMID: 20104212
- PMCID: PMC2862538
- DOI: 10.1038/mt.2009.316
HIV sequence variation associated with env antisense adoptive T-cell therapy in the hNSG mouse model
Abstract
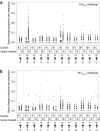
The first use of lentiviral vectors in humans involved transduction of mature T-cells with an human immunodeficiency virus (HIV)-derived env antisense (envAS) vector to protect cells from HIV infection. In that study, only a minority of the patient T-cell population could be gene-modified, raising the question of whether the altered cells could affect replicating HIV populations. We investigated this using humanized NOD/SCID IL-2Rgamma(null) (hNSG) mice reconstituted with approximately 4-11% envAS-modified human T-cells. Mice were challenged with HIV-1(NL4-3), which has an env perfectly complementary to envAS, or with HIV-1(BaL), which has a divergent env. No differences were seen in viral titer between mice that received envAS-modified cells and control mice that did not. Using 454/Roche pyrosequencing, we analyzed the mutational spectrum in HIV populations in serum-from 33 mice we recovered 84,074 total reads comprising 31,290 unique sequence variants. We found enrichment of A-to-G transitions and deletions in envAS-treated mice, paralleling a previous tissue culture study where most target cells contained envAS, even though minority of cells were envAS-modified here. Unexpectedly, this enrichment was only detected after the challenge with HIV-1(BaL), where the viral genome would form an imperfect duplex with envAS, and not HIV-1(NL4-3), where a perfectly matched duplex would form.
Figures

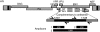

Similar articles
-
Enhanced inhibition of human immunodeficiency virus type 1 replication by novel lentiviral vectors expressing human immunodeficiency virus type 1 envelope antisense RNA.Hum Gene Ther. 2002 Jun 10;13(9):1027-37. doi: 10.1089/104303402753812430. Hum Gene Ther. 2002. PMID: 12067436
-
A combination anti-HIV-1 gene therapy approach using a single transcription unit that expresses antisense, decoy, and sense RNAs, and trans-dominant negative mutant Gag and Env proteins.Front Biosci. 2002 Feb 1;7:a15-28. doi: 10.2741/ding. Front Biosci. 2002. PMID: 11815282
-
Potent inhibition of human immunodeficiency virus type 1 replication by conditionally replicating human immunodeficiency virus-based lentiviral vectors expressing envelope antisense mRNA.Hum Gene Ther. 2000 Sep 20;11(14):2025-37. doi: 10.1089/10430340050143444. Hum Gene Ther. 2000. PMID: 11020801
-
The human HIV/peripheral blood lymphocyte (PBL)-SCID mouse. A modified human PBL-SCID model for the study of HIV pathogenesis and therapy.J Immunol. 1995 Jun 15;154(12):6612-23. J Immunol. 1995. PMID: 7759895
-
Anti-HIV-1 gene expressing lentiviral vectors as an adjunctive therapy for HIV-1 infection.Curr HIV Res. 2004 Apr;2(2):185-91. doi: 10.2174/1570162043484906. Curr HIV Res. 2004. PMID: 15078182 Review.
Cited by
-
Autonomous targeting of infectious superspreaders using engineered transmissible therapies.PLoS Comput Biol. 2011 Mar;7(3):e1002015. doi: 10.1371/journal.pcbi.1002015. Epub 2011 Mar 17. PLoS Comput Biol. 2011. PMID: 21483468 Free PMC article.
-
Supraphysiologic control over HIV-1 replication mediated by CD8 T cells expressing a re-engineered CD4-based chimeric antigen receptor.PLoS Pathog. 2017 Oct 12;13(10):e1006613. doi: 10.1371/journal.ppat.1006613. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 29023549 Free PMC article.
-
Survival of the fittest: positive selection of CD4+ T cells expressing a membrane-bound fusion inhibitor following HIV-1 infection.PLoS One. 2010 Aug 23;5(8):e12357. doi: 10.1371/journal.pone.0012357. PLoS One. 2010. PMID: 20808813 Free PMC article.
-
Potent and Broad Inhibition of HIV-1 by a Peptide from the gp41 Heptad Repeat-2 Domain Conjugated to the CXCR4 Amino Terminus.PLoS Pathog. 2016 Nov 17;12(11):e1005983. doi: 10.1371/journal.ppat.1005983. eCollection 2016 Nov. PLoS Pathog. 2016. PMID: 27855210 Free PMC article.
-
The utility of the new generation of humanized mice to study HIV-1 infection: transmission, prevention, pathogenesis, and treatment.Retrovirology. 2011 Aug 11;8:65. doi: 10.1186/1742-4690-8-65. Retrovirology. 2011. PMID: 21835012 Free PMC article. Review.
References
-
- Reyes-Darias JA, Sánchez-Luque FJ., and , Berzal-Herranz A. Inhibition of HIV-1 replication by RNA-based strategies. Curr HIV Res. 2008;6:500–514. - PubMed
-
- Scherer L, Rossi JJ., and , Weinberg MS. Progress and prospects: RNA-based therapies for treatment of HIV infection. Gene Ther. 2007;14:1057–1064. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

