The evolutionary fate of the genes encoding the purine catabolic enzymes in hominoids, birds, and reptiles
- PMID: 20106906
- PMCID: PMC2877996
- DOI: 10.1093/molbev/msq022
The evolutionary fate of the genes encoding the purine catabolic enzymes in hominoids, birds, and reptiles
Abstract
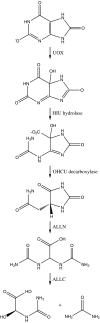
Gene loss has been proposed to play a major role in adaptive evolution, and recent studies are beginning to reveal its importance in human evolution. However, the potential consequence of a single gene-loss event upon the fates of functionally interrelated genes is poorly understood. Here, we use the purine metabolic pathway as a model system in which to explore this important question. The loss of urate oxidase (UOX) activity, a necessary step in this pathway, has occurred independently in the hominoid and bird/reptile lineages. Because the loss of UOX would have removed the functional constraint upon downstream genes in this pathway, these downstream genes are generally assumed to have subsequently deteriorated. In this study, we used a comparative genomics approach to empirically determine the fate of UOX itself and the downstream genes in five hominoids, two birds, and a reptile. Although we found that the loss of UOX likely triggered the genetic deterioration of the immediate downstream genes in the hominoids, surprisingly in the birds and reptiles, the UOX locus itself and some of the downstream genes were present in the genome and predicted to encode proteins. To account for the variable pattern of gene retention and loss after the inactivation of UOX, we hypothesize that although gene loss is a common fate for genes that have been rendered obsolete due to the upstream loss of an enzyme a metabolic pathway, it is also possible that same lack of constraint will foster the evolution of new functions or allow the optimization of preexisting alternative functions in the downstream genes, thereby resulting in gene retention. Thus, adaptive single-gene losses have the potential to influence the long-term evolutionary fate of functionally interrelated genes.
Figures


References
-
- Andersen O, Aas TS, Skugor S, Takle H, van Nes S, Grisdale-Helland B, Helland SJ, Terjesen BF. Purine-induced expression of urate oxidase and enzyme activity in Atlantic salmon (Salmo salar). Cloning of urate oxidase liver cDNA from three teleost species and the African lungfish Protopterus annectens. FEBS J. 2006;273:2839–2850. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

