Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis
- PMID: 20106958
- PMCID: PMC2822924
- DOI: 10.1261/rna.1852310
Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis
Erratum in
- RNA. 2010 Jul;16(7):1447
Abstract
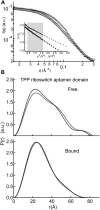
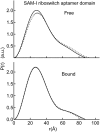
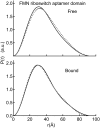
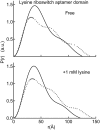
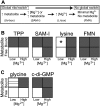
Riboswitches are structured mRNA elements that regulate gene expression upon binding specific cellular metabolites. It is thought that the highly conserved metabolite-binding domains of riboswitches undergo conformational change upon binding their cognate ligands. To investigate the generality of such a mechanism, we employed small-angle X-ray scattering (SAXS). We probed the nature of the global metabolite-induced response of the metabolite-binding domains of four different riboswitches that bind, respectively, thiamine pyrophosphate (TPP), flavin mononucleotide (FMN), lysine, and S-adenosyl methionine (SAM). We find that each RNA is unique in its global structural response to metabolite. Whereas some RNAs exhibit distinct free and bound conformations, others are globally insensitive to the presence of metabolite. Thus, a global conformational change of the metabolite-binding domain is not a requirement for riboswitch function. It is possible that the range of behaviors observed by SAXS, rather than being a biophysical idiosyncrasy, reflects adaptation of riboswitches to the regulatory requirements of their individual genomic context.
Figures
References
-
- Baird NJ, Westhof E, Qin H, Pan T, Sosnick TR. Structure of a folding intermediate reveals the interplay between core and peripheral elements in RNA folding. J Mol Biol. 2005;352:712–722. - PubMed
-
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. - PubMed
-
- Chauhan S, Caliskan G, Briber RM, Perez-Salas U, Rangan P, Thirumalai D, Woodson SA. RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme. J Mol Biol. 2005;353:1199–1209. - PubMed
-
- Dann CE, III, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. - PubMed
-
- Edwards TE, Ferré-D'Amaré AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
