Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms
- PMID: 20106967
- PMCID: PMC2856997
- DOI: 10.1074/jbc.M109.069179
Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms
Abstract
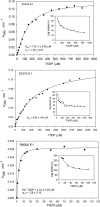
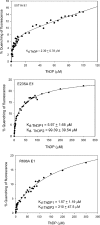
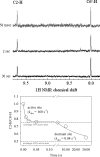
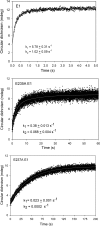
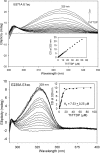
Kinetic, spectroscopic, and structural analysis tested the hypothesis that a chain of residues connecting the 4'-aminopyrimidine N1' atoms of thiamin diphosphates (ThDPs) in the two active centers of the Escherichia coli pyruvate dehydrogenase complex E1 component provides a signal transduction pathway. Substitution of the three acidic residues (Glu(571), Glu(235), and Glu(237)) and Arg(606) resulted in impaired binding of the second ThDP, once the first active center was filled, suggesting a pathway for communication between the two ThDPs. 1) Steady-state kinetic and fluorescence quenching studies revealed that upon E571A, E235A, E237A, and R606A substitutions, ThDP binding in the second active center was affected. 2) Analysis of the kinetics of thiazolium C2 hydrogen/deuterium exchange of enzyme-bound ThDP suggests half-of-the-sites reactivity for the E1 component, with fast (activated site) and slow exchanging sites (dormant site). The E235A and E571A variants gave no evidence for the slow exchanging site, indicating that only one of two active sites is filled with ThDP. 3) Titration of the E235A and E237A variants with methyl acetylphosphonate monitored by circular dichroism suggested that only half of the active sites were filled with a covalent predecarboxylation intermediate analog. 4) Crystal structures of E235A and E571A in complex with ThDP revealed the structural basis for the spectroscopic and kinetic observations and showed that either substitution affects cofactor binding, despite the fact that Glu(235) makes no direct contact with the cofactor. The role of the conserved Glu(571) residue in both catalysis and cofactor orientation is revealed by the combined results for the first time.
Figures




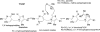



Similar articles
-
Glutamate 636 of the Escherichia coli pyruvate dehydrogenase-E1 participates in active center communication and behaves as an engineered acetolactate synthase with unusual stereoselectivity.J Biol Chem. 2005 Jun 3;280(22):21473-82. doi: 10.1074/jbc.M502691200. Epub 2005 Mar 31. J Biol Chem. 2005. PMID: 15802265
-
Structural determinants of enzyme binding affinity: the E1 component of pyruvate dehydrogenase from Escherichia coli in complex with the inhibitor thiamin thiazolone diphosphate.Biochemistry. 2004 Mar 9;43(9):2405-11. doi: 10.1021/bi030200y. Biochemistry. 2004. PMID: 14992577
-
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by thiamin 2-thiazolone and thiamin 2-thiothiazolone diphosphates. Evidence for reversible tight-binding inhibition.J Biol Chem. 2001 Dec 7;276(49):45969-78. doi: 10.1074/jbc.M104116200. Epub 2001 Oct 2. J Biol Chem. 2001. PMID: 11583990
-
Sixty years of thiamin diphosphate biochemistry.Biochim Biophys Acta. 1998 Jun 29;1385(2):177-86. doi: 10.1016/s0167-4838(98)00067-3. Biochim Biophys Acta. 1998. PMID: 9655906 Review.
-
Regulation of thiamin diphosphate-dependent 2-oxo acid decarboxylases by substrate and thiamin diphosphate.Mg(II) - evidence for tertiary and quaternary interactions.Biochim Biophys Acta. 1998 Jun 29;1385(2):287-306. doi: 10.1016/s0167-4838(98)00075-2. Biochim Biophys Acta. 1998. PMID: 9655921 Review.
Cited by
-
Potent Inhibition of E. coli DXP Synthase by a gem-Diaryl Bisubstrate Analog.ACS Infect Dis. 2024 Apr 12;10(4):1312-1326. doi: 10.1021/acsinfecdis.3c00734. Epub 2024 Mar 21. ACS Infect Dis. 2024. PMID: 38513073 Free PMC article.
-
Novel insights into transketolase activation by cofactor binding identifies two native species subpopulations.Sci Rep. 2019 Nov 6;9(1):16116. doi: 10.1038/s41598-019-52647-y. Sci Rep. 2019. PMID: 31695144 Free PMC article.
-
The pyruvate dehydrogenase complexes: structure-based function and regulation.J Biol Chem. 2014 Jun 13;289(24):16615-23. doi: 10.1074/jbc.R114.563148. Epub 2014 May 5. J Biol Chem. 2014. PMID: 24798336 Free PMC article. Review.
-
Structure of the native pyruvate dehydrogenase complex reveals the mechanism of substrate insertion.Nat Commun. 2021 Sep 6;12(1):5277. doi: 10.1038/s41467-021-25570-y. Nat Commun. 2021. PMID: 34489474 Free PMC article.
-
Glyoxylate carboligase: a unique thiamin diphosphate-dependent enzyme that can cycle between the 4'-aminopyrimidinium and 1',4'-iminopyrimidine tautomeric forms in the absence of the conserved glutamate.Biochemistry. 2012 Oct 9;51(40):7940-52. doi: 10.1021/bi300893v. Epub 2012 Sep 25. Biochemistry. 2012. PMID: 22970650 Free PMC article.
References
-
- Jordan F., Nemeria N. S., Sergienko E. (2005) Acc. Chem. Res. 38, 755–763 - PubMed
-
- Frank R. A., Titman C. M., Pratap J. V., Luisi B. F., Perham R. N. (2004) Science 306, 872–876 - PubMed
-
- Jordan F. (2004) Science 306, 818–820 - PubMed
-
- Ciszak E. M., Korotchkina L. G., Dominiak P. M., Sidhu S., Patel M. S. (2003) J. Biol. Chem. 278, 21240–21246 - PubMed
-
- Seifert F., Golbik R., Brauer J., Lilie H., Schröder-Tittmann K., Hinze E., Korotchkina L. G., Patel M. S., Tittmann K. (2006) Biochemistry 45, 12775–12785 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

