Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase
- PMID: 20106986
- PMCID: PMC2859556
- DOI: 10.1074/jbc.M109.068908
Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase
Abstract
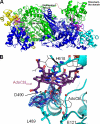
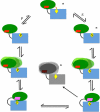
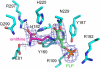
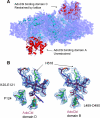
D-ornithine 4,5-aminomutase (OAM) from Clostridium sticklandii converts D-ornithine to 2,4-diaminopentanoic acid by way of radical propagation from an adenosylcobalamin (AdoCbl) to a pyridoxal 5'-phosphate (PLP) cofactor. We have solved OAM crystal structures in different catalytic states that together demonstrate unusual stability of the AdoCbl Co-C bond and that radical catalysis is coupled to large-scale domain motion. The 2.0-A substrate-free enzyme crystal structure reveals the Rossmann domain, harboring the intact AdoCbl cofactor, is tilted toward the edge of the PLP binding triose-phosphate isomerase barrel domain. The PLP forms an internal aldimine link to the Rossmann domain through Lys(629), effectively locking the enzyme in this "open" pre-catalytic conformation. The distance between PLP and 5'-deoxyadenosyl group is 23 A, and large-scale domain movement is thus required prior to radical catalysis. The OAM crystals contain two Rossmann domains within the asymmetric unit that are unconstrained by the crystal lattice. Surprisingly, the binding of various ligands to OAM crystals (in an oxygen-free environment) leads to transimination in the absence of significant reorientation of the Rossmann domains. In contrast, when performed under aerobic conditions, this leads to extreme disorder in the latter domains correlated with the loss of the 5'-deoxyadenosyl group. Our data indicate turnover and hence formation of the "closed" conformation is occurring within OAM crystals, but that the equilibrium is poised toward the open conformation. We propose that substrate binding induces large-scale domain motion concomitant with a reconfiguration of the 5'-deoxyadenosyl group, triggering radical catalysis in OAM.
Figures

Similar articles
-
Large-scale domain motions and pyridoxal-5'-phosphate assisted radical catalysis in coenzyme B12-dependent aminomutases.Int J Mol Sci. 2014 Feb 20;15(2):3064-87. doi: 10.3390/ijms15023064. Int J Mol Sci. 2014. PMID: 24562332 Free PMC article. Review.
-
A conformational sampling model for radical catalysis in pyridoxal phosphate- and cobalamin-dependent enzymes.J Biol Chem. 2014 Dec 5;289(49):34161-74. doi: 10.1074/jbc.M114.590471. Epub 2014 Sep 11. J Biol Chem. 2014. PMID: 25213862 Free PMC article.
-
A locking mechanism preventing radical damage in the absence of substrate, as revealed by the x-ray structure of lysine 5,6-aminomutase.Proc Natl Acad Sci U S A. 2004 Nov 9;101(45):15870-5. doi: 10.1073/pnas.0407074101. Epub 2004 Oct 28. Proc Natl Acad Sci U S A. 2004. PMID: 15514022 Free PMC article.
-
Mechanism of radical-based catalysis in the reaction catalyzed by adenosylcobalamin-dependent ornithine 4,5-aminomutase.J Biol Chem. 2008 Dec 12;283(50):34615-25. doi: 10.1074/jbc.M807911200. Epub 2008 Oct 22. J Biol Chem. 2008. PMID: 18948256 Free PMC article.
-
Adenosylcobalamin enzymes: theory and experiment begin to converge.Biochim Biophys Acta. 2012 Nov;1824(11):1154-64. doi: 10.1016/j.bbapap.2012.03.012. Epub 2012 Apr 3. Biochim Biophys Acta. 2012. PMID: 22516318 Free PMC article. Review.
Cited by
-
A mechanochemical switch to control radical intermediates.Biochemistry. 2014 Jun 17;53(23):3830-8. doi: 10.1021/bi500050k. Epub 2014 Jun 6. Biochemistry. 2014. PMID: 24846280 Free PMC article.
-
Cofactor Selectivity in Methylmalonyl Coenzyme A Mutase, a Model Cobamide-Dependent Enzyme.mBio. 2019 Sep 24;10(5):e01303-19. doi: 10.1128/mBio.01303-19. mBio. 2019. PMID: 31551329 Free PMC article.
-
Large-scale domain motions and pyridoxal-5'-phosphate assisted radical catalysis in coenzyme B12-dependent aminomutases.Int J Mol Sci. 2014 Feb 20;15(2):3064-87. doi: 10.3390/ijms15023064. Int J Mol Sci. 2014. PMID: 24562332 Free PMC article. Review.
-
Orchestrated Domain Movement in Catalysis by Cytochrome P450 Reductase.Sci Rep. 2017 Aug 29;7(1):9741. doi: 10.1038/s41598-017-09840-8. Sci Rep. 2017. PMID: 28852004 Free PMC article.
-
Identification of a Novel Cobamide Remodeling Enzyme in the Beneficial Human Gut Bacterium Akkermansia muciniphila.mBio. 2020 Dec 8;11(6):e02507-20. doi: 10.1128/mBio.02507-20. mBio. 2020. PMID: 33293380 Free PMC article.
References
-
- Henzler-Wildman K. A., Lei M., Thai V., Kerns S. J., Karplus M., Kern D. (2007) Nature 450, 913–916 - PubMed
-
- Boehr D. D., Dyson H. J., Wright P. E. (2006) Chem. Rev. 106, 3055–3079 - PubMed
-
- Barker H. A. (1981) Annu. Rev. Biochem. 50, 23–40 - PubMed
-
- Chen H. P., Wu S. H., Lin Y. L., Chen C. M., Tsay S. S. (2001) J. Biol. Chem. 276, 44744–44750 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

