The HR motif in the RNA-dependent RNA polymerase L protein of Chandipura virus is required for unconventional mRNA-capping activity
- PMID: 20107017
- PMCID: PMC2855774
- DOI: 10.1099/vir.0.019307-0
The HR motif in the RNA-dependent RNA polymerase L protein of Chandipura virus is required for unconventional mRNA-capping activity
Abstract
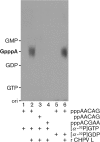
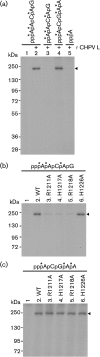
Chandipura virus (CHPV) is an emerging human pathogen associated with acute encephalitis and is related closely to vesicular stomatitis virus (VSV), a prototype rhabdovirus. Here, we demonstrate that the RNA polymerase L protein of CHPV exhibits a VSV-like RNA:GDP polyribonucleotidyltransferase (PRNTase) activity, which transfers the 5'-monophosphorylated (p-) viral mRNA start sequence to GDP to produce a capped RNA, and that the conserved HR motif in the CHPV L protein is essential for the PRNTase activity. Interestingly, the CHPV L protein was found to form two distinct SDS-resistant complexes with the CHPV mRNA and leader RNA start sequences; mutations in the HR motif significantly reduced the formation of the former complex (a putative covalent enzyme-pRNA intermediate in the PRNTase reaction), but not the latter complex. These results suggest that the rhabdoviral L proteins universally use the active-site HR motif for the PRNTase reaction at the step of the enzyme-pRNA intermediate formation.
Figures
References
-
- Bourhy, H., Kissi, B. & Tordo, N. (1993). Molecular diversity of the Lyssavirus genus. Virology 194, 70–81. - PubMed
-
- Chadha, M. S., Arankalle, V. A., Jadi, R. S., Joshi, M. V., Thakare, J. P., Mahadev, P. V. & Mishra, A. C. (2005). An outbreak of Chandipura virus encephalitis in the eastern districts of Gujarat state, India. Am J Trop Med Hyg 73, 566–570. - PubMed
-
- Hoffmann, B., Schutze, H. & Mettenleiter, T. C. (2002). Determination of the complete genomic sequence and analysis of the gene products of the virus of spring viremia of carp, a fish rhabdovirus. Virus Res 84, 89–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

