The timing and location of glial cell line-derived neurotrophic factor expression determine enteric nervous system structure and function
- PMID: 20107080
- PMCID: PMC2836725
- DOI: 10.1523/JNEUROSCI.3861-09.2010
The timing and location of glial cell line-derived neurotrophic factor expression determine enteric nervous system structure and function
Abstract
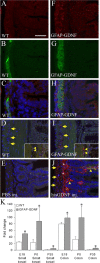
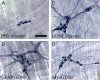
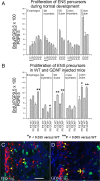
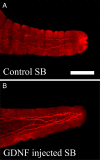
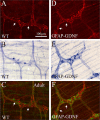
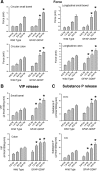
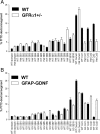
Ret signaling is critical for formation of the enteric nervous system (ENS) because Ret activation promotes ENS precursor survival, proliferation, and migration and provides trophic support for mature enteric neurons. Although these roles are well established, we now provide evidence that increasing levels of the Ret ligand glial cell line-derived neurotrophic factor (GDNF) in mice causes alterations in ENS structure and function that are critically dependent on the time and location of increased GDNF availability. This is demonstrated using two different strains of transgenic mice and by injecting newborn mice with GDNF. Furthermore, because different subclasses of ENS precursors withdraw from the cell cycle at different times during development, increases in GDNF at specific times alter the ratio of neuronal subclasses in the mature ENS. In addition, we confirm that esophageal neurons are GDNF responsive and demonstrate that the location of GDNF production influences neuronal process projection for NADPH diaphorase-expressing, but not acetylcholinesterase-, choline acetyltransferase-, or tryptophan hydroxylase-expressing, small bowel myenteric neurons. We further demonstrate that changes in GDNF availability influence intestinal function in vitro and in vivo. Thus, changes in GDNF expression can create a wide variety of alterations in ENS structure and function and may in part contribute to human motility disorders.
Figures



References
-
- Airaksinen MS, Saarma M. The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–394. - PubMed
-
- Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G, Borrego S, Pelet A, Arnold S, Miao X, Griseri P, Brooks AS, Antinolo G, de Pontual L, Clement-Ziza M, Munnich A, Kashuk C, West K, Wong KK, Lyonnet S, Chakravarti A, Tam PK, Ceccherini I, Hofstra RM, Fernandez R. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14. - PubMed
-
- Baloh RH, Enomoto H, Johnson EM, Jr, Milbrandt J. The GDNF family ligands and receptors: implications for neural development. Curr Opin Neurobiol. 2000;10:103–110. - PubMed
-
- Blaugrund E, Pham TD, Tennyson VM, Lo L, Sommer L, Anderson DJ, Gershon MD. Distinct subpopulations of enteric neuronal progenitors defined by time of development, sympathoadrenal lineage markers, and Mash-1-dependence. Development. 1996;122:309–320. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
