Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection
- PMID: 20107183
- PMCID: PMC2863030
- DOI: 10.4049/jimmunol.0903704
Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection
Abstract
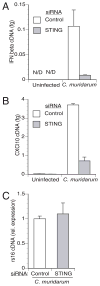
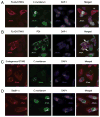
Type I IFN signaling has recently been shown to be detrimental to the host during infection with Chlamydia muridarum in both mouse lung and female genital tract. However, the pattern recognition receptor and the signaling pathways involved in chlamydial-induced IFN-beta are unclear. Previous studies have demonstrated no role for TLR4 and a partial role for MyD88 in chlamydial-induced IFN-beta. In this study, we demonstrate that mouse macrophages lacking TLR3, TRIF, TLR7, or TLR9 individually or both TLR4 and MyD88, still induce IFN-beta equivalent to wild type controls, leading to the hypothesis that TLR-independent cytosolic pathogen receptor pathways are crucial for this response. Silencing nucleotide-binding oligomerization domain 1 in HeLa cells partially decreased chlamydial-induced IFN-beta. Independently, small interfering RNA-mediated knockdown of the stimulator of IFN gene (STING) protein in HeLa cells and mouse oviduct epithelial cells significantly decreased IFN-beta mRNA expression, suggesting a critical role for STING in chlamydial-induced IFN-beta induction. Conversely, silencing of mitochondria-associated antiviral signaling proteins and the Rig-I-like receptors, RIG-I, and melanoma differentiation associated protein 5, had no effect. In addition, induction of IFN-beta depended on the downstream transcription IFN regulatory factor 3, and on activation of NF-kappaB and MAPK p38. Finally, STING, an endoplasmic reticulum-resident protein, was found to localize in close proximity to the chlamydial inclusion membrane during infection. These results indicate that C. muridarum induces IFN-beta via stimulation of nucleotide-binding oligomerization domain 1 pathway, and TLR- and Rig-I-like receptor-independent pathways that require STING, culminating in activation of IFN regulatory factor 3, NF-kappaB, and p38 MAPK.
Conflict of interest statement
The authors have no financial conflicts of interest.
Figures




References
-
- Xia M, Bumgarner RE, Lampe MF, Stamm WE. Chlamydia trachomatis infection alters host cell transcription in diverse cellular pathways. J Infect Dis. 2003;187:424–434. - PubMed
-
- Rasmussen SJ, Eckmann L, Quayle AJ, Shen L, Zhang YX, Anderson DJ, Fierer J, Stephens RS, Kagnoff MF. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J Clin Invest. 1997;99:77–87. - PMC - PubMed
-
- Ren Q, Robertson SJ, Howe D, Barrows LF, Heinzen RA. Comparative DNA microarray analysis of host cell transcriptional responses to infection by Coxiella burnetii or Chlamydia trachomatis. Ann N Y Acad Sci. 2003;990:701–713. - PubMed
-
- Lad SP, Fukuda EY, Li J, de la Maza LM, Li E. Up-regulation of the JAK/STAT1 signal pathway during Chlamydia trachomatis infection. J Immunol. 2005;174:7186–7193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

