Copy neutral loss of heterozygosity: a novel chromosomal lesion in myeloid malignancies
- PMID: 20107230
- PMCID: PMC2854422
- DOI: 10.1182/blood-2009-10-201848
Copy neutral loss of heterozygosity: a novel chromosomal lesion in myeloid malignancies
Abstract
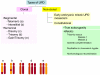
Single nucleotide polymorphism arrays (SNP-A) have recently been widely applied as a powerful karyotyping tool in numerous translational cancer studies. SNP-A complements traditional metaphase cytogenetics with the unique ability to delineate a previously hidden chromosomal defect, copy neutral loss of heterozygosity (CN-LOH). Emerging data demonstrate that selected hematologic malignancies exhibit abundant CN-LOH, often in the setting of a normal metaphase karyotype and no previously identified clonal marker. In this review, we explore emerging biologic and clinical features of CN-LOH relevant to hematologic malignancies. In myeloid malignancies, CN-LOH has been associated with the duplication of oncogenic mutations with concomitant loss of the normal allele. Examples include JAK2, MPL, c-KIT, and FLT3. More recent investigations have focused on evaluation of candidate genes contained in common CN-LOH and deletion regions and have led to the discovery of tumor suppressor genes, including c-CBL and family members, as well as TET2. Investigations into the underlying mechanisms generating CN-LOH have great promise for elucidating general cancer mechanisms. We anticipate that further detailed characterization of CN-LOH lesions will probably facilitate our discovery of a more complete set of pathogenic molecular lesions, disease and prognosis markers, and better understanding of the initiation and progression of hematologic malignancies.
Figures
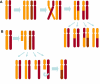



References
-
- Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. - PubMed
-
- Kuiper RP, Schoenmakers EF, van Reijmersdal SV, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21(6):1258–1266. - PubMed
-
- Sulong S, Moorman AV, Irving JA, et al. A comprehensive analysis of the CDKN2A gene in childhood acute lymphoblastic leukemia reveals genomic deletion, copy number neutral loss of heterozygosity, and association with specific cytogenetic subgroups. Blood. 2009;113(1):100–107. - PubMed
-
- Irving JA, Bloodworth L, Bown NP, Case MC, Hogarth LA, Hall AG. Loss of heterozygosity in childhood acute lymphoblastic leukemia detected by genome-wide microarray single nucleotide polymorphism analysis. Cancer Res. 2005;65(8):3053–3058. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

