Dynamic regulation of genes involved in mitochondrial DNA replication and transcription during mouse brown fat cell differentiation and recruitment
- PMID: 20107496
- PMCID: PMC2809086
- DOI: 10.1371/journal.pone.0008458
Dynamic regulation of genes involved in mitochondrial DNA replication and transcription during mouse brown fat cell differentiation and recruitment
Abstract
Background: Brown adipocytes are specialised in dissipating energy through adaptive thermogenesis, whereas white adipocytes are specialised in energy storage. These essentially opposite functions are possible for two reasons relating to mitochondria, namely expression of uncoupling protein 1 (UCP1) and a remarkably higher mitochondrial abundance in brown adipocytes.
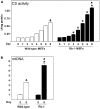
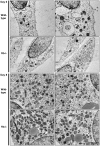
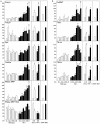
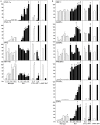
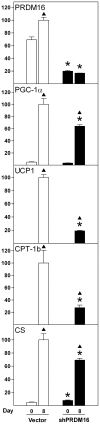
Methodology/principal findings: Here we report a comprehensive characterisation of gene expression linked to mitochondrial DNA replication, transcription and function during white and brown fat cell differentiation in vitro as well as in white and brown fat, brown adipose tissue fractions and in selected adipose tissues during cold exposure. We find a massive induction of the majority of such genes during brown adipocyte differentiation and recruitment, e.g. of the mitochondrial transcription factors A (Tfam) and B2 (Tfb2m), whereas only a subset of the same genes were induced during white adipose conversion. In addition, PR domain containing 16 (PRDM16) was found to be expressed at substantially higher levels in brown compared to white pre-adipocytes and adipocytes. We demonstrate that forced expression of Tfam but not Tfb2m in brown adipocyte precursor cells promotes mitochondrial DNA replication, and that silencing of PRDM16 expression during brown fat cell differentiation blunts mitochondrial biogenesis and expression of brown fat cell markers.
Conclusions/significance: Using both in vitro and in vivo model systems of white and brown fat cell differentiation, we report a detailed characterisation of gene expression linked to mitochondrial biogenesis and function. We find significant differences in differentiating white and brown adipocytes, which might explain the notable increase in mitochondrial content observed during brown adipose conversion. In addition, our data support a key role of PRDM16 in triggering brown adipocyte differentiation, including mitochondrial biogenesis and expression of UCP1.
Conflict of interest statement
Figures



Similar articles
-
Stat5a promotes brown adipocyte differentiation and thermogenic program through binding and transactivating the Kdm6a promoter.Cell Cycle. 2020 Apr;19(8):895-905. doi: 10.1080/15384101.2020.1731644. Epub 2020 Mar 24. Cell Cycle. 2020. PMID: 32207362 Free PMC article.
-
The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation.Am J Physiol Endocrinol Metab. 2010 Jun;298(6):E1244-53. doi: 10.1152/ajpendo.00600.2009. Epub 2010 Mar 30. Am J Physiol Endocrinol Metab. 2010. PMID: 20354155
-
Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes.J Biol Chem. 2010 Mar 5;285(10):7153-64. doi: 10.1074/jbc.M109.053942. Epub 2009 Dec 22. J Biol Chem. 2010. PMID: 20028987 Free PMC article.
-
UCP1 protein: The molecular hub of adipose organ plasticity.Biochimie. 2017 Mar;134:71-76. doi: 10.1016/j.biochi.2016.09.008. Epub 2016 Sep 10. Biochimie. 2017. PMID: 27622583 Review.
-
Meaningful respirometric measurements of UCP1-mediated thermogenesis.Biochimie. 2017 Mar;134:56-61. doi: 10.1016/j.biochi.2016.12.005. Epub 2016 Dec 14. Biochimie. 2017. PMID: 27986537 Review.
Cited by
-
Effects of Fatty Acid Metabolites on Adipocytes Britening: Role of Thromboxane A2.Cells. 2023 Jan 30;12(3):446. doi: 10.3390/cells12030446. Cells. 2023. PMID: 36766790 Free PMC article.
-
GSK3 is a negative regulator of the thermogenic program in brown adipocytes.Sci Rep. 2018 Feb 22;8(1):3469. doi: 10.1038/s41598-018-21795-y. Sci Rep. 2018. PMID: 29472592 Free PMC article.
-
Chronic cold exposure induces autophagy to promote fatty acid oxidation, mitochondrial turnover, and thermogenesis in brown adipose tissue.iScience. 2021 Apr 15;24(5):102434. doi: 10.1016/j.isci.2021.102434. eCollection 2021 May 21. iScience. 2021. PMID: 34027318 Free PMC article.
-
miR-125b affects mitochondrial biogenesis and impairs brite adipocyte formation and function.Mol Metab. 2016 Jun 15;5(8):615-625. doi: 10.1016/j.molmet.2016.06.005. eCollection 2016 Aug. Mol Metab. 2016. PMID: 27656399 Free PMC article.
-
DACRA induces profound weight loss, satiety control, and increased mitochondrial respiratory capacity in adipose tissue.Int J Obes (Lond). 2024 Oct;48(10):1421-1429. doi: 10.1038/s41366-024-01564-w. Epub 2024 Jun 15. Int J Obes (Lond). 2024. PMID: 38879729
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

