Evolutionary mirages: selection on binding site composition creates the illusion of conserved grammars in Drosophila enhancers
- PMID: 20107516
- PMCID: PMC2809757
- DOI: 10.1371/journal.pgen.1000829
Evolutionary mirages: selection on binding site composition creates the illusion of conserved grammars in Drosophila enhancers
Abstract
The clustering of transcription factor binding sites in developmental enhancers and the apparent preferential conservation of clustered sites have been widely interpreted as proof that spatially constrained physical interactions between transcription factors are required for regulatory function. However, we show here that selection on the composition of enhancers alone, and not their internal structure, leads to the accumulation of clustered sites with evolutionary dynamics that suggest they are preferentially conserved. We simulated the evolution of idealized enhancers from Drosophila melanogaster constrained to contain only a minimum number of binding sites for one or more factors. Under this constraint, mutations that destroy an existing binding site are tolerated only if a compensating site has emerged elsewhere in the enhancer. Overlapping sites, such as those frequently observed for the activator Bicoid and repressor Krüppel, had significantly longer evolutionary half-lives than isolated sites for the same factors. This leads to a substantially higher density of overlapping sites than expected by chance and the appearance that such sites are preferentially conserved. Because D. melanogaster (like many other species) has a bias for deletions over insertions, sites tended to become closer together over time, leading to an overall clustering of sites in the absence of any selection for clustered sites. Since this effect is strongest for the oldest sites, clustered sites also incorrectly appear to be preferentially conserved. Following speciation, sites tend to be closer together in all descendent species than in their common ancestors, violating the common assumption that shared features of species' genomes reflect their ancestral state. Finally, we show that selection on binding site composition alone recapitulates the observed number of overlapping and closely neighboring sites in real D. melanogaster enhancers. Thus, this study calls into question the common practice of inferring "cis-regulatory grammars" from the organization and evolutionary dynamics of developmental enhancers.
Conflict of interest statement
MBE is a co-founder of the Public Library of Science and a member of its Board of Directors.
Figures

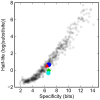
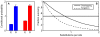

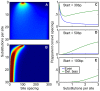

Comment in
-
Illusions of conservation.Nat Rev Genet. 2010 Mar;11(3):169. doi: 10.1038/nrg2753. Nat Rev Genet. 2010. PMID: 21485426 No abstract available.
Similar articles
-
Molecular dissection of cis-regulatory modules at the Drosophila bithorax complex reveals critical transcription factor signature motifs.Dev Biol. 2011 Nov 15;359(2):290-302. doi: 10.1016/j.ydbio.2011.07.028. Epub 2011 Jul 28. Dev Biol. 2011. PMID: 21821017 Free PMC article.
-
Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change.Development. 1998 Mar;125(5):949-58. doi: 10.1242/dev.125.5.949. Development. 1998. PMID: 9449677
-
Evolutionary origins of transcription factor binding site clusters.Mol Biol Evol. 2012 Mar;29(3):1059-70. doi: 10.1093/molbev/msr277. Epub 2011 Nov 10. Mol Biol Evol. 2012. PMID: 22075113 Free PMC article.
-
One thousand and one ways of making functionally similar transcriptional enhancers.Bioessays. 2008 Nov;30(11-12):1052-7. doi: 10.1002/bies.20849. Bioessays. 2008. PMID: 18937349 Review.
-
Transcriptional Enhancers in Drosophila.Genetics. 2020 Sep;216(1):1-26. doi: 10.1534/genetics.120.301370. Genetics. 2020. PMID: 32878914 Free PMC article. Review.
Cited by
-
Mapping the complexity of transcription control in higher eukaryotes.Genome Biol. 2010;11(4):115. doi: 10.1186/gb-2010-11-4-115. Epub 2010 Apr 30. Genome Biol. 2010. PMID: 20441601 Free PMC article.
-
Neutral forces acting on intragenomic variability shape the Escherichia coli regulatory network topology.Proc Natl Acad Sci U S A. 2013 May 7;110(19):7754-9. doi: 10.1073/pnas.1217630110. Epub 2013 Apr 22. Proc Natl Acad Sci U S A. 2013. PMID: 23610404 Free PMC article.
-
Simulations of enhancer evolution provide mechanistic insights into gene regulation.Mol Biol Evol. 2014 Jan;31(1):184-200. doi: 10.1093/molbev/mst170. Epub 2013 Oct 4. Mol Biol Evol. 2014. PMID: 24097306 Free PMC article.
-
Mechanisms of mutational robustness in transcriptional regulation.Front Genet. 2015 Oct 27;6:322. doi: 10.3389/fgene.2015.00322. eCollection 2015. Front Genet. 2015. PMID: 26579194 Free PMC article. Review.
-
Global reference mapping of human transcription factor footprints.Nature. 2020 Jul;583(7818):729-736. doi: 10.1038/s41586-020-2528-x. Epub 2020 Jul 29. Nature. 2020. PMID: 32728250 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

