Nonsense-mediated decay enables intron gain in Drosophila
- PMID: 20107520
- PMCID: PMC2809761
- DOI: 10.1371/journal.pgen.1000819
Nonsense-mediated decay enables intron gain in Drosophila
Abstract
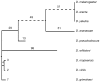
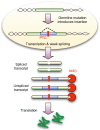
Intron number varies considerably among genomes, but despite their fundamental importance, the mutational mechanisms and evolutionary processes underlying the expansion of intron number remain unknown. Here we show that Drosophila, in contrast to most eukaryotic lineages, is still undergoing a dramatic rate of intron gain. These novel introns carry significantly weaker splice sites that may impede their identification by the spliceosome. Novel introns are more likely to encode a premature termination codon (PTC), indicating that nonsense-mediated decay (NMD) functions as a backup for weak splicing of new introns. Our data suggest that new introns originate when genomic insertions with weak splice sites are hidden from selection by NMD. This mechanism reduces the sequence requirement imposed on novel introns and implies that the capacity of the spliceosome to recognize weak splice sites was a prerequisite for intron gain during eukaryotic evolution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Detection and analysis of alternative splicing in Yarrowia lipolytica reveal structural constraints facilitating nonsense-mediated decay of intron-retaining transcripts.Genome Biol. 2010;11(6):R65. doi: 10.1186/gb-2010-11-6-r65. Epub 2010 Jun 23. Genome Biol. 2010. PMID: 20573210 Free PMC article.
-
Origin and evolution of spliceosomal introns.Biol Direct. 2012 Apr 16;7:11. doi: 10.1186/1745-6150-7-11. Biol Direct. 2012. PMID: 22507701 Free PMC article. Review.
-
A spliceosomal intron in Giardia lamblia.Proc Natl Acad Sci U S A. 2002 Mar 19;99(6):3701-5. doi: 10.1073/pnas.042700299. Epub 2002 Feb 19. Proc Natl Acad Sci U S A. 2002. PMID: 11854456 Free PMC article.
-
Selection on Position of Nonsense Codons in Introns.Genetics. 2016 Nov;204(3):1239-1248. doi: 10.1534/genetics.116.189894. Epub 2016 Sep 14. Genetics. 2016. PMID: 27630196 Free PMC article.
-
Coevolution of genomic intron number and splice sites.Trends Genet. 2007 Jul;23(7):321-5. doi: 10.1016/j.tig.2007.04.001. Epub 2007 Apr 18. Trends Genet. 2007. PMID: 17442445 Review.
Cited by
-
Phylogenetic distribution of intron positions in alpha-amylase genes of bilateria suggests numerous gains and losses.PLoS One. 2011;6(5):e19673. doi: 10.1371/journal.pone.0019673. Epub 2011 May 17. PLoS One. 2011. PMID: 21611157 Free PMC article.
-
Intron creation and DNA repair.Cell Mol Life Sci. 2011 Jan;68(2):235-42. doi: 10.1007/s00018-010-0532-2. Epub 2010 Sep 19. Cell Mol Life Sci. 2011. PMID: 20853128 Free PMC article. Review.
-
Some novel intron positions in conserved Drosophila genes are caused by intron sliding or tandem duplication.BMC Evol Biol. 2010 May 26;10:156. doi: 10.1186/1471-2148-10-156. BMC Evol Biol. 2010. PMID: 20500887 Free PMC article.
-
Quantitative RNA-seq meta-analysis of alternative exon usage in C. elegans.Genome Res. 2017 Dec;27(12):2120-2128. doi: 10.1101/gr.224626.117. Epub 2017 Oct 31. Genome Res. 2017. PMID: 29089372 Free PMC article.
-
Molecular trajectories leading to the alternative fates of duplicate genes.PLoS One. 2012;7(6):e38958. doi: 10.1371/journal.pone.0038958. Epub 2012 Jun 14. PLoS One. 2012. PMID: 22720000 Free PMC article.
References
-
- Coulombe-Huntington J, Majewski J. Intron Loss and Gain in Drosophila. Molecular Biology and Evolution. 2007;24:2842–2850. - PubMed
-
- Irimia M, Rukov JL, Penny D, Vinther J, Garcia-Fernandez J, et al. Origin of introns by ‘intronization’ of exonic sequences. Trends Genet. 2008;24:378–381. - PubMed
-
- Cavalier-Smith T. Intron phylogeny: a new hypothesis. Trends Genet. 1991;7:145–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

