TRIM5alpha Modulates Immunodeficiency Virus Control in Rhesus Monkeys
- PMID: 20107597
- PMCID: PMC2809762
- DOI: 10.1371/journal.ppat.1000738
TRIM5alpha Modulates Immunodeficiency Virus Control in Rhesus Monkeys
Abstract
The cytoplasmic TRIM5alpha proteins of certain mammalian lineages efficiently recognize the incoming capsids of particular retroviruses and potently restrict infection in a species-specific manner. Successful retroviruses have evolved capsids that are less efficiently recognized by the TRIM5alpha proteins of the natural hosts. To address whether TRIM5alpha contributes to the outcome of retroviral infection in a susceptible host species, we investigated the impact of TRIM5 polymorphisms in rhesus monkeys on the course of a simian immunodeficiency virus (SIV) infection. Full-length TRIM5alpha cDNAs were derived from each of 79 outbred monkeys and sequenced. Associations were explored between the expression of particular TRIM5 alleles and both the permissiveness of cells to SIV infection in vitro and clinical sequelae of SIV infection in vivo. Natural variation in the TRIM5alpha B30.2(SPRY) domain influenced the efficiency of SIVmac capsid binding and the in vitro susceptibility of cells from the monkeys to SIVmac infection. We also show the importance in vivo of the interaction of SIVmac with different allelic forms of TRIM5, demonstrating that particular alleles are associated with as much as 1.3 median log difference in set-point viral loads in SIVmac-infected rhesus monkeys. Moreover, these allelic forms of TRIM5 were associated with the extent of loss of central memory (CM) CD4+ T cells and the rate of progression to AIDS in the infected monkeys. These findings demonstrate a central role for TRIM5alpha in limiting the replication of an immunodeficiency virus infection in a primate host.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
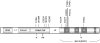

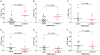



Similar articles
-
Gain-of-sensitivity mutations in a Trim5-resistant primary isolate of pathogenic SIV identify two independent conserved determinants of Trim5α specificity.PLoS Pathog. 2013 May;9(5):e1003352. doi: 10.1371/journal.ppat.1003352. Epub 2013 May 9. PLoS Pathog. 2013. PMID: 23675300 Free PMC article.
-
TRIM5α does not affect simian immunodeficiency virus SIV(mac251) replication in vaccinated or unvaccinated Indian rhesus macaques following intrarectal challenge exposure.J Virol. 2011 Dec;85(23):12399-409. doi: 10.1128/JVI.05707-11. Epub 2011 Sep 14. J Virol. 2011. PMID: 21917950 Free PMC article.
-
Differential restriction of human immunodeficiency virus type 2 and simian immunodeficiency virus SIVmac by TRIM5alpha alleles.J Virol. 2005 Sep;79(18):11580-7. doi: 10.1128/JVI.79.18.11580-11587.2005. J Virol. 2005. PMID: 16140735 Free PMC article.
-
TRIM5 alpha drives SIVsmm evolution in rhesus macaques.PLoS Pathog. 2013;9(8):e1003577. doi: 10.1371/journal.ppat.1003577. Epub 2013 Aug 22. PLoS Pathog. 2013. PMID: 23990789 Free PMC article.
-
Anti-retroviral activity of TRIM5 alpha.Rev Med Virol. 2010 Mar;20(2):77-92. doi: 10.1002/rmv.637. Rev Med Virol. 2010. PMID: 20049904 Review.
Cited by
-
Immune and Genetic Correlates of Vaccine Protection Against Mucosal Infection by SIV in Monkeys.Sci Transl Med. 2011 May 4;3(81):81ra36. doi: 10.1126/scitranslmed.3002351. Sci Transl Med. 2011. PMID: 21543722 Free PMC article.
-
Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV.Nature. 2014 Jan 23;505(7484):502-8. doi: 10.1038/nature12893. Epub 2013 Dec 18. Nature. 2014. PMID: 24352234 Free PMC article.
-
Vaccine-induced immune responses against both Gag and Env improve control of simian immunodeficiency virus replication in rectally challenged rhesus macaques.PLoS Pathog. 2017 Jul 21;13(7):e1006529. doi: 10.1371/journal.ppat.1006529. eCollection 2017 Jul. PLoS Pathog. 2017. PMID: 28732035 Free PMC article.
-
A trivalent recombinant Ad5 gag/pol/nef vaccine fails to protect rhesus macaques from infection or control virus replication after a limiting-dose heterologous SIV challenge.Vaccine. 2012 Jun 22;30(30):4465-75. doi: 10.1016/j.vaccine.2012.04.082. Epub 2012 May 6. Vaccine. 2012. PMID: 22569124 Free PMC article.
-
Immunization with wild-type or CD4-binding-defective HIV-1 Env trimers reduces viremia equivalently following heterologous challenge with simian-human immunodeficiency virus.J Virol. 2010 Sep;84(18):9086-95. doi: 10.1128/JVI.01015-10. Epub 2010 Jul 7. J Virol. 2010. PMID: 20610729 Free PMC article.
References
-
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

