Mechanism and specificity of a symmetrical benzimidazolephenylcarboxamide helicase inhibitor
- PMID: 20108979
- PMCID: PMC2832472
- DOI: 10.1021/bi901974a
Mechanism and specificity of a symmetrical benzimidazolephenylcarboxamide helicase inhibitor
Abstract
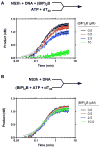
This study examines the effects of 1-N,4-N-bis[4-(1H-benzimidazol-2-yl)phenyl]benzene-1,4-dicarboxamide ((BIP)(2)B) on the NS3 helicase encoded by the hepatitis C virus (HCV). Molecular beacon-based helicase assays were used to show that (BIP)(2)B inhibits the ability of HCV helicase to separate a variety of RNA and DNA duplexes with half-maximal inhibitory concentrations ranging from 0.7 to 5 microM, depending on the nature of the substrate. In single turnover assays, (BIP)(2)B only inhibited unwinding reactions when it was preincubated with the helicase-nucleic acid complex. (BIP)(2)B quenched NS3 intrinsic protein fluorescence with an apparent dissociation constant of 5 microM, and in the presence of (BIP)(2)B, HCV helicase did not appear to interact with a fluorescent DNA oligonucleotide. In assays monitoring HCV helicase-catalyzed ATP hydrolysis, (BIP)(2)B only inhibited helicase-catalyzed ATP hydrolysis in the presence of intermediate concentrations of RNA, suggesting RNA and (BIP)(2)B compete for the same binding site. HCV helicases isolated from various HCV genotypes were similarly sensitive to (BIP)(2)B, with half-maximal inhibitory concentrations ranging from 0.7 to 2.4 microM. (BIP)(2)B also inhibited ATP hydrolysis catalyzed by related helicases from Dengue virus, Japanese encephalitis virus, and humans. (BIP)(2)B appeared to bind the HCV and human proteins with similar affinity (K(i) = 7 and 8 microM, respectively), but it bound the flavivirus proteins up to 270 times more tightly. Results are discussed in light of a molecular model of a (BIP)(2)B-HCV helicase complex, which is unable to bind nucleic acid, thus preventing the enzyme from separating double-stranded nucleic acid.
Figures
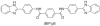
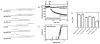




References
-
- Phoon CW, Ng PY, Ting AE, Yeo SL, Sim MM. Biological evaluation of hepatitis C virus helicase inhibitors. Bioorg Med Chem Lett. 2001;11:1647–1650. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

