A novel scoring function for discriminating hyperthermophilic and mesophilic proteins with application to predicting relative thermostability of protein mutants
- PMID: 20109199
- PMCID: PMC3098108
- DOI: 10.1186/1471-2105-11-62
A novel scoring function for discriminating hyperthermophilic and mesophilic proteins with application to predicting relative thermostability of protein mutants
Abstract
Background: The ability to design thermostable proteins is theoretically important and practically useful. Robust and accurate algorithms, however, remain elusive. One critical problem is the lack of reliable methods to estimate the relative thermostability of possible mutants.
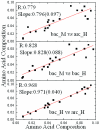
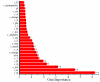
Results: We report a novel scoring function for discriminating hyperthermophilic and mesophilic proteins with application to predicting the relative thermostability of protein mutants. The scoring function was developed based on an elaborate analysis of a set of features calculated or predicted from 540 pairs of hyperthermophilic and mesophilic protein ortholog sequences. It was constructed by a linear combination of ten important features identified by a feature ranking procedure based on the random forest classification algorithm. The weights of these features in the scoring function were fitted by a hill-climbing algorithm. This scoring function has shown an excellent ability to discriminate hyperthermophilic from mesophilic sequences. The prediction accuracies reached 98.9% and 97.3% in discriminating orthologous pairs in training and the holdout testing datasets, respectively. Moreover, the scoring function can distinguish non-homologous sequences with an accuracy of 88.4%. Additional blind tests using two datasets of experimentally investigated mutations demonstrated that the scoring function can be used to predict the relative thermostability of proteins and their mutants at very high accuracies (92.9% and 94.4%). We also developed an amino acid substitution preference matrix between mesophilic and hyperthermophilic proteins, which may be useful in designing more thermostable proteins.
Conclusions: We have presented a novel scoring function which can distinguish not only HP/MP ortholog pairs, but also non-homologous pairs at high accuracies. Most importantly, it can be used to accurately predict the relative stability of proteins and their mutants, as demonstrated in two blind tests. In addition, the residue substitution preference matrix assembled in this study may reflect the thermal adaptation induced substitution biases. A web server implementing the scoring function and the dataset used in this study are freely available at http://www.abl.ku.edu/thermorank/.
Figures


Similar articles
-
Predicting protein thermostability changes from sequence upon multiple mutations.Bioinformatics. 2008 Jul 1;24(13):i190-5. doi: 10.1093/bioinformatics/btn166. Bioinformatics. 2008. PMID: 18586713 Free PMC article.
-
PRBP: Prediction of RNA-Binding Proteins Using a Random Forest Algorithm Combined with an RNA-Binding Residue Predictor.IEEE/ACM Trans Comput Biol Bioinform. 2015 Nov-Dec;12(6):1385-93. doi: 10.1109/TCBB.2015.2418773. IEEE/ACM Trans Comput Biol Bioinform. 2015. PMID: 26671809
-
[Random forest for classification of thermophilic and psychrophilic proteins based on amino acid composition distribution].Sheng Wu Gong Cheng Xue Bao. 2008 Feb;24(2):302-8. Sheng Wu Gong Cheng Xue Bao. 2008. PMID: 18464617 Chinese.
-
Different packing of external residues can explain differences in the thermostability of proteins from thermophilic and mesophilic organisms.Bioinformatics. 2007 Sep 1;23(17):2231-8. doi: 10.1093/bioinformatics/btm345. Epub 2007 Jun 28. Bioinformatics. 2007. PMID: 17599925
-
Protein thermostability: structure-based difference of amino acid between thermophilic and mesophilic proteins.J Biotechnol. 2004 Aug 5;111(3):269-77. doi: 10.1016/j.jbiotec.2004.01.018. J Biotechnol. 2004. PMID: 15246663
Cited by
-
Mapping QTL for the traits associated with heat tolerance in wheat (Triticum aestivum L.).BMC Genet. 2014 Nov 11;15:97. doi: 10.1186/s12863-014-0097-4. BMC Genet. 2014. PMID: 25384418 Free PMC article.
-
Novel Ricin Subunit Antigens With Enhanced Capacity to Elicit Toxin-Neutralizing Antibody Responses in Mice.J Pharm Sci. 2016 May;105(5):1603-1613. doi: 10.1016/j.xphs.2016.02.009. Epub 2016 Mar 15. J Pharm Sci. 2016. PMID: 26987947 Free PMC article.
-
Prediction and design of thermostable proteins with a desired melting temperature.Sci Rep. 2025 May 14;15(1):16683. doi: 10.1038/s41598-025-98667-9. Sci Rep. 2025. PMID: 40369176 Free PMC article.
-
PROTS-RF: a robust model for predicting mutation-induced protein stability changes.PLoS One. 2012;7(10):e47247. doi: 10.1371/journal.pone.0047247. Epub 2012 Oct 15. PLoS One. 2012. PMID: 23077576 Free PMC article.
-
The use of consensus sequence information to engineer stability and activity in proteins.Methods Enzymol. 2020;643:149-179. doi: 10.1016/bs.mie.2020.06.001. Epub 2020 Jul 17. Methods Enzymol. 2020. PMID: 32896279 Free PMC article.
References
-
- Schweiker KL, Makhatadze GI. A Computational Approach for the Rational Design of Stable Proteins and Enzymes: Optimization of Surface Charge-Charge Interactions. Methods in Enzymology: Computer Methods. 2009;454(Pt A):175–211. full_text. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

