Characterization of X-linked SNP genotypic variation in globally distributed human populations
- PMID: 20109212
- PMCID: PMC2847713
- DOI: 10.1186/gb-2010-11-1-r10
Characterization of X-linked SNP genotypic variation in globally distributed human populations
Abstract
Background: The transmission pattern of the human X chromosome reduces its population size relative to the autosomes, subjects it to disproportionate influence by female demography, and leaves X-linked mutations exposed to selection in males. As a result, the analysis of X-linked genomic variation can provide insights into the influence of demography and selection on the human genome. Here we characterize the genomic variation represented by 16,297 X-linked SNPs genotyped in the CEPH human genome diversity project samples.
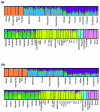
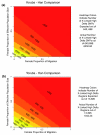
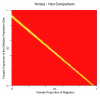
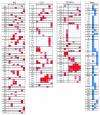
Results: We found that X chromosomes tend to be more differentiated between human populations than autosomes, with several notable exceptions. Comparisons between genetically distant populations also showed an excess of X-linked SNPs with large allele frequency differences. Combining information about these SNPs with results from tests designed to detect selective sweeps, we identified two regions that were clear outliers from the rest of the X chromosome for haplotype structure and allele frequency distribution. We were also able to more precisely define the geographical extent of some previously described X-linked selective sweeps.
Conclusions: The relationship between male and female demographic histories is likely to be complex as evidence supporting different conclusions can be found in the same dataset. Although demography may have contributed to the excess of SNPs with large allele frequency differences observed on the X chromosome, we believe that selection is at least partially responsible. Finally, our results reveal the geographical complexities of selective sweeps on the X chromosome and argue for the use of diverse populations in studies of selection.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

