ICC-CLASS: isotopically-coded cleavable crosslinking analysis software suite
- PMID: 20109223
- PMCID: PMC2827373
- DOI: 10.1186/1471-2105-11-64
ICC-CLASS: isotopically-coded cleavable crosslinking analysis software suite
Abstract
Background: Successful application of crosslinking combined with mass spectrometry for studying proteins and protein complexes requires specifically-designed crosslinking reagents, experimental techniques, and data analysis software. Using isotopically-coded ("heavy and light") versions of the crosslinker and cleavable crosslinking reagents is analytically advantageous for mass spectrometric applications and provides a "handle" that can be used to distinguish crosslinked peptides of different types, and to increase the confidence of the identification of the crosslinks.
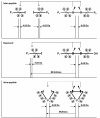
Results: Here, we describe a program suite designed for the analysis of mass spectrometric data obtained with isotopically-coded cleavable crosslinkers. The suite contains three programs called: DX, DXDX, and DXMSMS. DX searches the mass spectra for the presence of ion signal doublets resulting from the light and heavy isotopic forms of the isotopically-coded crosslinking reagent used. DXDX searches for possible mass matches between cleaved and uncleaved isotopically-coded crosslinks based on the established chemistry of the cleavage reaction for a given crosslinking reagent. DXMSMS assigns the crosslinks to the known protein sequences, based on the isotopically-coded and un-coded MS/MS fragmentation data of uncleaved and cleaved peptide crosslinks.
Conclusion: The combination of these three programs, which are tailored to the analytical features of the specific isotopically-coded cleavable crosslinking reagents used, represents a powerful software tool for automated high-accuracy peptide crosslink identification. See: http://www.creativemolecules.com/CM_Software.htm.
Figures



References
-
- Bennett KL, Kussmann M, Björk P, Godzwon M, Mikkelsen M, Sørensen P, Roepstorff P. Chemical cross-linking with thiol-cleavable reagents combined with differential mass spectrometric peptide mapping--a novel approach to assess intermolecular protein contacts. Protein Sci. 2000;9:1503–18. doi: 10.1110/ps.9.8.1503. - DOI - PMC - PubMed
-
- ProteinProspector. http://prospector.ucsf.edu
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

