Haplotype allelic classes for detecting ongoing positive selection
- PMID: 20109229
- PMCID: PMC2831848
- DOI: 10.1186/1471-2105-11-65
Haplotype allelic classes for detecting ongoing positive selection
Abstract
Background: Natural selection eliminates detrimental and favors advantageous phenotypes. This process leaves characteristic signatures in underlying genomic segments that can be recognized through deviations in allelic or haplotypic frequency spectra. To provide an identifiable signature of recent positive selection that can be detected by comparison with the background distribution, we introduced a new way of looking at genomic polymorphisms: haplotype allelic classes.
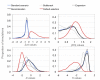
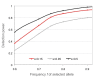
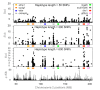
Results: The model combines segregating sites and haplotypic information in order to reveal useful data characteristics. We developed a summary statistic, Svd, to compare the distribution of the haplotypes carrying the selected allele with the distribution of the remaining ones. Coalescence simulations are used to study the distributions under standard population models assuming neutrality, demographic scenarios and selection models. To test, in practice, haplotype allelic class performance and the derived statistic in capturing deviation from neutrality due to positive selection, we analyzed haplotypic variation in detail in the locus of lactase persistence in the three HapMap Phase II populations.
Conclusions: We showed that the Svd statistic is less sensitive than other tests to confounding factors such as demography or recombination. Our approach succeeds in identifying candidate loci, such as the lactase-persistence locus, as targets of strong positive selection and provides a new tool complementary to other tests to study natural selection in genomic data.
Figures



References
-
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, Ackerman HC, Campbell SJ, Altshuler D, Cooper R, Kwiatkowski D, Ward R, Lander ES. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–837. doi: 10.1038/nature01140. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

